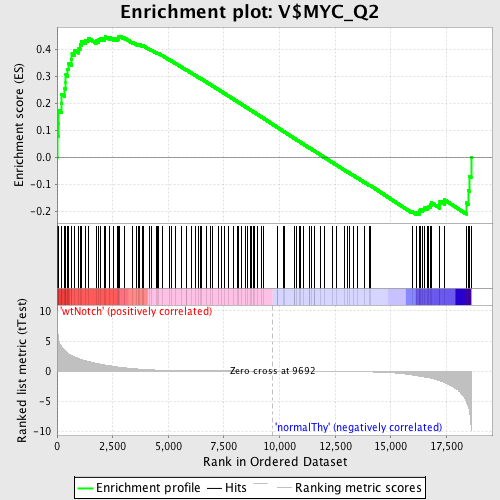
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy_repos |
| Phenotype | phenotype_wtNotch_versus_normalThy.cls#wtNotch_versus_normalThy_repos |
| Upregulated in class | wtNotch |
| GeneSet | V$MYC_Q2 |
| Enrichment Score (ES) | 0.44790733 |
| Normalized Enrichment Score (NES) | 1.2096773 |
| Nominal p-value | 0.087628864 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TIMM10 | 610070 1450019 | 3 | 9.579 | 0.0793 | Yes | ||
| 2 | ATAD3A | 4050288 | 44 | 5.911 | 0.1262 | Yes | ||
| 3 | TAGLN2 | 4560121 | 49 | 5.773 | 0.1738 | Yes | ||
| 4 | PA2G4 | 2030463 | 194 | 4.069 | 0.1998 | Yes | ||
| 5 | LDHA | 2190594 | 205 | 4.012 | 0.2325 | Yes | ||
| 6 | PFDN2 | 4230156 | 348 | 3.411 | 0.2532 | Yes | ||
| 7 | SHMT2 | 1850180 6040010 | 392 | 3.256 | 0.2778 | Yes | ||
| 8 | NUDC | 4780025 | 395 | 3.235 | 0.3046 | Yes | ||
| 9 | HNRPA1 | 4670398 5050687 | 481 | 2.980 | 0.3247 | Yes | ||
| 10 | CFL1 | 2340735 | 498 | 2.954 | 0.3483 | Yes | ||
| 11 | EIF4B | 5390563 5270577 | 653 | 2.610 | 0.3617 | Yes | ||
| 12 | PPRC1 | 1740441 | 668 | 2.584 | 0.3823 | Yes | ||
| 13 | HMGN2 | 3140091 | 779 | 2.385 | 0.3962 | Yes | ||
| 14 | FKBP11 | 3170575 | 979 | 2.067 | 0.4026 | Yes | ||
| 15 | GPD1 | 2480095 | 1032 | 1.991 | 0.4163 | Yes | ||
| 16 | COPS7A | 3940458 7000022 | 1085 | 1.933 | 0.4295 | Yes | ||
| 17 | ADK | 380338 520180 5270524 | 1283 | 1.731 | 0.4332 | Yes | ||
| 18 | RPL22 | 4070070 | 1401 | 1.628 | 0.4404 | Yes | ||
| 19 | KRTCAP2 | 5220040 | 1759 | 1.298 | 0.4319 | Yes | ||
| 20 | BCL9L | 6940053 | 1877 | 1.210 | 0.4356 | Yes | ||
| 21 | AK2 | 4590458 4760195 6110097 | 1965 | 1.135 | 0.4403 | Yes | ||
| 22 | TAF6L | 1850056 | 2149 | 1.008 | 0.4387 | Yes | ||
| 23 | PABPC4 | 1990170 6760270 5390138 | 2166 | 1.001 | 0.4462 | Yes | ||
| 24 | IVNS1ABP | 4760601 6520113 | 2343 | 0.915 | 0.4442 | Yes | ||
| 25 | ATP5F1 | 7000725 | 2551 | 0.787 | 0.4396 | Yes | ||
| 26 | STMN1 | 1990717 | 2698 | 0.711 | 0.4376 | Yes | ||
| 27 | PSEN2 | 130382 | 2754 | 0.678 | 0.4402 | Yes | ||
| 28 | GTF2H1 | 2570746 2650148 | 2773 | 0.670 | 0.4448 | Yes | ||
| 29 | CDC14A | 4050132 | 2817 | 0.652 | 0.4479 | Yes | ||
| 30 | B4GALT2 | 780037 | 3016 | 0.565 | 0.4419 | No | ||
| 31 | FABP3 | 6860452 | 3384 | 0.436 | 0.4257 | No | ||
| 32 | CNNM1 | 2190433 | 3583 | 0.365 | 0.4180 | No | ||
| 33 | IL15RA | 2570270 3190711 6100091 6100270 | 3677 | 0.330 | 0.4157 | No | ||
| 34 | AMPD2 | 630093 1170070 | 3701 | 0.323 | 0.4171 | No | ||
| 35 | CTBP2 | 430309 3710079 | 3854 | 0.287 | 0.4113 | No | ||
| 36 | COMMD3 | 2510259 | 3862 | 0.285 | 0.4133 | No | ||
| 37 | TIAL1 | 4150048 6510605 | 4148 | 0.228 | 0.3998 | No | ||
| 38 | UBXD3 | 5050193 | 4242 | 0.212 | 0.3965 | No | ||
| 39 | AP3M1 | 5340433 | 4468 | 0.182 | 0.3859 | No | ||
| 40 | RAB3IL1 | 380685 5420176 | 4532 | 0.176 | 0.3839 | No | ||
| 41 | EBNA1BP2 | 50369 1400433 6040019 | 4561 | 0.173 | 0.3838 | No | ||
| 42 | ALDH3B1 | 4210010 6940403 | 4722 | 0.156 | 0.3765 | No | ||
| 43 | ILK | 870601 4480180 | 5035 | 0.129 | 0.3607 | No | ||
| 44 | CHD4 | 5420059 6130338 6380717 | 5129 | 0.123 | 0.3567 | No | ||
| 45 | TESK2 | 6520524 | 5325 | 0.111 | 0.3471 | No | ||
| 46 | SYT6 | 2510280 3850128 4540064 | 5583 | 0.098 | 0.3340 | No | ||
| 47 | PITX3 | 3130162 | 5797 | 0.087 | 0.3232 | No | ||
| 48 | RFX5 | 5420128 | 5806 | 0.086 | 0.3235 | No | ||
| 49 | RUSC1 | 6450093 6980575 | 6058 | 0.076 | 0.3105 | No | ||
| 50 | COL2A1 | 460446 3940500 | 6227 | 0.070 | 0.3020 | No | ||
| 51 | AGMAT | 6200154 | 6368 | 0.065 | 0.2950 | No | ||
| 52 | SC5DL | 6650390 | 6447 | 0.062 | 0.2913 | No | ||
| 53 | LTBR | 2450341 | 6473 | 0.061 | 0.2905 | No | ||
| 54 | SLC1A7 | 1990181 | 6693 | 0.055 | 0.2791 | No | ||
| 55 | RTN4RL2 | 6450609 | 6876 | 0.050 | 0.2697 | No | ||
| 56 | EIF3S10 | 1190079 3840176 | 6975 | 0.048 | 0.2648 | No | ||
| 57 | USP15 | 610592 3520504 | 7252 | 0.042 | 0.2502 | No | ||
| 58 | PLA2G4A | 6380364 | 7405 | 0.039 | 0.2423 | No | ||
| 59 | LIN28 | 6590672 | 7537 | 0.036 | 0.2355 | No | ||
| 60 | RHEBL1 | 2810576 | 7702 | 0.033 | 0.2269 | No | ||
| 61 | POGK | 5290315 | 7921 | 0.029 | 0.2154 | No | ||
| 62 | HOXC12 | 5270411 | 8100 | 0.026 | 0.2060 | No | ||
| 63 | HOXC13 | 4070722 | 8173 | 0.024 | 0.2023 | No | ||
| 64 | TFB2M | 3830161 4670059 4760184 | 8288 | 0.022 | 0.1963 | No | ||
| 65 | NRAS | 6900577 | 8445 | 0.019 | 0.1880 | No | ||
| 66 | CGN | 5890139 6370167 6400537 | 8549 | 0.018 | 0.1826 | No | ||
| 67 | BHLHB3 | 3450438 | 8696 | 0.015 | 0.1748 | No | ||
| 68 | NR0B2 | 2470692 | 8755 | 0.014 | 0.1718 | No | ||
| 69 | TRIM46 | 3520609 4590010 | 8827 | 0.013 | 0.1681 | No | ||
| 70 | B3GALT6 | 50195 | 8861 | 0.013 | 0.1664 | No | ||
| 71 | B3GALT2 | 430136 | 9017 | 0.010 | 0.1581 | No | ||
| 72 | HNRPF | 670138 | 9176 | 0.008 | 0.1496 | No | ||
| 73 | CHRM1 | 4280619 | 9255 | 0.007 | 0.1455 | No | ||
| 74 | CTSF | 6100021 | 9893 | -0.003 | 0.1111 | No | ||
| 75 | LRP8 | 3610746 5360035 | 9894 | -0.003 | 0.1111 | No | ||
| 76 | SIRT1 | 1190731 | 10156 | -0.007 | 0.0970 | No | ||
| 77 | SLC16A1 | 50433 | 10203 | -0.007 | 0.0946 | No | ||
| 78 | ALX4 | 3870019 | 10659 | -0.014 | 0.0701 | No | ||
| 79 | IPO7 | 2190746 | 10682 | -0.015 | 0.0691 | No | ||
| 80 | NKX2-3 | 1990736 6940088 | 10781 | -0.016 | 0.0639 | No | ||
| 81 | LHX9 | 2350112 | 10874 | -0.018 | 0.0591 | No | ||
| 82 | FADS3 | 3120079 | 10921 | -0.018 | 0.0567 | No | ||
| 83 | OPRD1 | 4280138 | 11066 | -0.021 | 0.0491 | No | ||
| 84 | NRIP3 | 1340372 | 11336 | -0.025 | 0.0348 | No | ||
| 85 | FOXD3 | 6550156 | 11359 | -0.026 | 0.0338 | No | ||
| 86 | APOA5 | 6130471 | 11412 | -0.027 | 0.0312 | No | ||
| 87 | PAX6 | 1190025 | 11552 | -0.029 | 0.0240 | No | ||
| 88 | SLC6A15 | 5290176 6040017 | 11562 | -0.029 | 0.0237 | No | ||
| 89 | HPCA | 6860010 | 11860 | -0.036 | 0.0080 | No | ||
| 90 | FOSL1 | 430021 | 11997 | -0.038 | 0.0009 | No | ||
| 91 | SFXN2 | 2650563 3140095 3830762 3870398 | 12373 | -0.047 | -0.0190 | No | ||
| 92 | TDRD1 | 4230301 6110750 | 12545 | -0.052 | -0.0278 | No | ||
| 93 | IPO13 | 3870088 | 12934 | -0.064 | -0.0482 | No | ||
| 94 | WEE1 | 3390070 | 13037 | -0.067 | -0.0532 | No | ||
| 95 | HPCAL4 | 2600484 | 13162 | -0.072 | -0.0593 | No | ||
| 96 | POU3F1 | 3710022 | 13330 | -0.080 | -0.0677 | No | ||
| 97 | CBX5 | 3830072 6290167 | 13506 | -0.088 | -0.0764 | No | ||
| 98 | ZZZ3 | 780133 2320239 2640452 5080706 6550341 | 13821 | -0.108 | -0.0925 | No | ||
| 99 | LMX1A | 2810239 5690324 | 14024 | -0.122 | -0.1024 | No | ||
| 100 | TGFB2 | 4920292 | 14066 | -0.126 | -0.1035 | No | ||
| 101 | SCYL1 | 5050717 | 15951 | -0.629 | -0.2002 | No | ||
| 102 | ATP6V0B | 2810112 | 16145 | -0.735 | -0.2045 | No | ||
| 103 | PFKFB3 | 630706 | 16270 | -0.815 | -0.2044 | No | ||
| 104 | RRAGC | 5910348 | 16277 | -0.821 | -0.1980 | No | ||
| 105 | FXYD2 | 1400280 2350719 3520184 7040377 | 16316 | -0.848 | -0.1930 | No | ||
| 106 | PTPRF | 1770528 3190044 | 16425 | -0.912 | -0.1912 | No | ||
| 107 | CSDA | 2470092 3060053 4780056 | 16492 | -0.958 | -0.1869 | No | ||
| 108 | ESRRA | 6550242 | 16633 | -1.031 | -0.1859 | No | ||
| 109 | CENTB5 | 5690128 | 16710 | -1.093 | -0.1809 | No | ||
| 110 | USP2 | 1190292 1240253 4850035 | 16789 | -1.162 | -0.1755 | No | ||
| 111 | H3F3A | 2900086 | 16833 | -1.191 | -0.1680 | No | ||
| 112 | FCHSD2 | 5720092 | 17198 | -1.593 | -0.1744 | No | ||
| 113 | ARL3 | 5690725 | 17204 | -1.603 | -0.1614 | No | ||
| 114 | ATF7IP | 2690176 6770021 | 17420 | -1.898 | -0.1573 | No | ||
| 115 | TMEM24 | 5860039 | 18389 | -4.948 | -0.1685 | No | ||
| 116 | RORC | 1740121 | 18500 | -6.120 | -0.1237 | No | ||
| 117 | FXYD6 | 3870164 | 18533 | -6.645 | -0.0703 | No | ||
| 118 | NIT1 | 1230338 3940671 | 18607 | -9.013 | 0.0005 | No |

