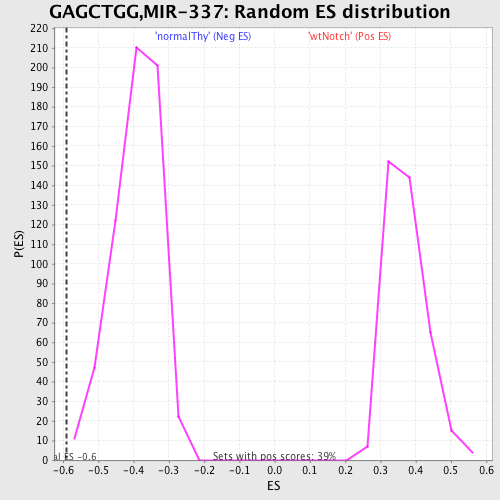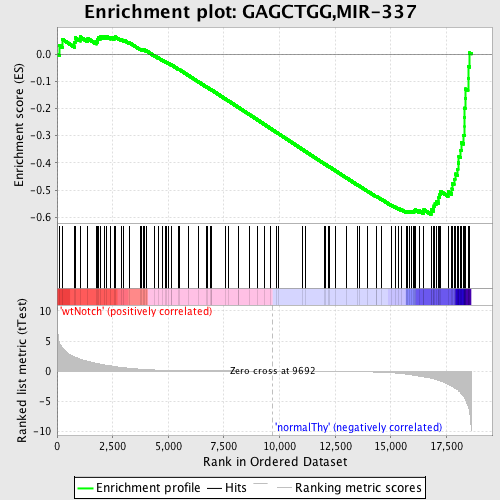
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy_repos |
| Phenotype | phenotype_wtNotch_versus_normalThy.cls#wtNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | GAGCTGG,MIR-337 |
| Enrichment Score (ES) | -0.5892677 |
| Normalized Enrichment Score (NES) | -1.5088669 |
| Nominal p-value | 0.004893964 |
| FDR q-value | 0.13280205 |
| FWER p-Value | 0.81 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PDGFRB | 6450279 | 97 | 4.773 | 0.0316 | No | ||
| 2 | BCAP29 | 2350500 | 232 | 3.876 | 0.0543 | No | ||
| 3 | EI24 | 3120433 | 781 | 2.381 | 0.0431 | No | ||
| 4 | ABCF1 | 2630707 2760273 3520725 | 812 | 2.333 | 0.0595 | No | ||
| 5 | GPD1 | 2480095 | 1032 | 1.991 | 0.0631 | No | ||
| 6 | MCRS1 | 110121 2340592 5270484 | 1383 | 1.640 | 0.0568 | No | ||
| 7 | DPH2 | 2100402 3060039 | 1756 | 1.300 | 0.0468 | No | ||
| 8 | H1F0 | 1400408 | 1826 | 1.248 | 0.0527 | No | ||
| 9 | RAB15 | 430070 | 1850 | 1.235 | 0.0610 | No | ||
| 10 | TOR2A | 1400026 | 1964 | 1.135 | 0.0637 | No | ||
| 11 | ACOT7 | 730735 1400176 7050053 | 2120 | 1.020 | 0.0632 | No | ||
| 12 | DGKZ | 3460041 6400129 | 2216 | 0.990 | 0.0657 | No | ||
| 13 | PARP6 | 3290332 4760167 6900181 | 2415 | 0.873 | 0.0617 | No | ||
| 14 | RNF19 | 6400315 | 2561 | 0.782 | 0.0599 | No | ||
| 15 | CAMK2G | 630097 | 2613 | 0.751 | 0.0630 | No | ||
| 16 | MRPS16 | 2510390 | 2892 | 0.621 | 0.0528 | No | ||
| 17 | IER5L | 2450154 | 3001 | 0.574 | 0.0514 | No | ||
| 18 | B3GAT3 | 1580438 | 3255 | 0.478 | 0.0414 | No | ||
| 19 | TWIST1 | 5130619 | 3727 | 0.315 | 0.0184 | No | ||
| 20 | SSBP3 | 870717 1050377 7040551 | 3805 | 0.298 | 0.0165 | No | ||
| 21 | COMMD3 | 2510259 | 3862 | 0.285 | 0.0157 | No | ||
| 22 | THRA | 6770341 110164 1990600 | 3910 | 0.274 | 0.0153 | No | ||
| 23 | TRIM35 | 2690603 | 4006 | 0.251 | 0.0121 | No | ||
| 24 | NFASC | 1940619 2470156 4590168 6840524 | 4397 | 0.192 | -0.0075 | No | ||
| 25 | BNIPL | 360139 5860022 | 4540 | 0.175 | -0.0138 | No | ||
| 26 | EFNB1 | 2970433 | 4725 | 0.156 | -0.0226 | No | ||
| 27 | VCP | 5340168 | 4860 | 0.144 | -0.0287 | No | ||
| 28 | MFN1 | 5080546 6200292 | 4926 | 0.138 | -0.0311 | No | ||
| 29 | SEMA6A | 2690138 3060021 | 4985 | 0.132 | -0.0333 | No | ||
| 30 | CHD4 | 5420059 6130338 6380717 | 5129 | 0.123 | -0.0400 | No | ||
| 31 | NR1D1 | 2360471 770746 6590204 | 5147 | 0.122 | -0.0400 | No | ||
| 32 | DSCAM | 1780050 2450731 2810438 | 5471 | 0.103 | -0.0567 | No | ||
| 33 | LRRC4 | 3850041 | 5506 | 0.101 | -0.0577 | No | ||
| 34 | RASGRP4 | 540102 | 5888 | 0.083 | -0.0776 | No | ||
| 35 | MAMDC1 | 3710129 4480075 | 6361 | 0.066 | -0.1026 | No | ||
| 36 | RNF121 | 110079 1580279 | 6717 | 0.054 | -0.1214 | No | ||
| 37 | KCNH8 | 3940129 | 6758 | 0.053 | -0.1232 | No | ||
| 38 | MAP3K9 | 3290707 | 6885 | 0.050 | -0.1296 | No | ||
| 39 | POGZ | 870348 7040286 | 6923 | 0.049 | -0.1312 | No | ||
| 40 | ABCA9 | 2760136 5550706 | 6942 | 0.049 | -0.1318 | No | ||
| 41 | KLF11 | 6840739 | 7586 | 0.035 | -0.1662 | No | ||
| 42 | SLC12A5 | 1980692 | 7692 | 0.033 | -0.1717 | No | ||
| 43 | SEPT6 | 1740739 5220402 | 8167 | 0.024 | -0.1971 | No | ||
| 44 | PACSIN2 | 70204 | 8171 | 0.024 | -0.1971 | No | ||
| 45 | CHRD | 3140368 | 8648 | 0.016 | -0.2227 | No | ||
| 46 | EHMT2 | 1410278 6900601 | 9016 | 0.010 | -0.2424 | No | ||
| 47 | LBX1 | 1300164 4060204 | 9302 | 0.006 | -0.2578 | No | ||
| 48 | ZMYND11 | 630181 2570019 4850138 5360040 | 9589 | 0.002 | -0.2732 | No | ||
| 49 | ITGA11 | 1740112 6590110 | 9849 | -0.002 | -0.2872 | No | ||
| 50 | KCND1 | 6860278 | 9944 | -0.003 | -0.2922 | No | ||
| 51 | SP7 | 1050315 | 11007 | -0.020 | -0.3494 | No | ||
| 52 | SLC1A2 | 2230520 | 11148 | -0.022 | -0.3568 | No | ||
| 53 | FOSL1 | 430021 | 11997 | -0.038 | -0.4024 | No | ||
| 54 | CD44 | 3990072 4850671 5860411 6860148 7050551 | 12081 | -0.040 | -0.4065 | No | ||
| 55 | BNC2 | 4810603 | 12219 | -0.044 | -0.4136 | No | ||
| 56 | AP1S3 | 510594 1450242 2190576 | 12233 | -0.044 | -0.4140 | No | ||
| 57 | HOXB5 | 2900538 | 12496 | -0.050 | -0.4277 | No | ||
| 58 | TEAD3 | 2320142 | 13026 | -0.067 | -0.4558 | No | ||
| 59 | KLF12 | 1660095 4810288 5340546 6520286 | 13492 | -0.087 | -0.4802 | No | ||
| 60 | ALDH1A2 | 2320301 | 13599 | -0.093 | -0.4852 | No | ||
| 61 | OVOL1 | 1690131 | 13948 | -0.117 | -0.5031 | No | ||
| 62 | NAV1 | 4210519 6620129 | 14351 | -0.154 | -0.5237 | No | ||
| 63 | JPH1 | 610739 | 14363 | -0.155 | -0.5231 | No | ||
| 64 | BTG2 | 2350411 | 14579 | -0.176 | -0.5333 | No | ||
| 65 | FOS | 1850315 | 15011 | -0.260 | -0.5546 | No | ||
| 66 | STIP1 | 6450390 | 15187 | -0.301 | -0.5617 | No | ||
| 67 | TYSND1 | 2940273 | 15361 | -0.356 | -0.5683 | No | ||
| 68 | SLC25A22 | 2360050 | 15490 | -0.404 | -0.5721 | No | ||
| 69 | GLCCI1 | 110091 2640082 2900692 3290070 6370014 | 15693 | -0.493 | -0.5792 | No | ||
| 70 | SORT1 | 1850242 2320632 3870537 | 15744 | -0.520 | -0.5779 | No | ||
| 71 | VAMP1 | 450735 1940707 | 15818 | -0.549 | -0.5776 | No | ||
| 72 | SFRS1 | 2360440 | 15931 | -0.616 | -0.5789 | No | ||
| 73 | GRK6 | 1500053 | 16014 | -0.659 | -0.5782 | No | ||
| 74 | ROM1 | 3130685 | 16051 | -0.681 | -0.5749 | No | ||
| 75 | FRAG1 | 1050504 1170152 2030092 | 16098 | -0.710 | -0.5719 | No | ||
| 76 | MAT2A | 4070026 4730079 6020280 | 16267 | -0.813 | -0.5747 | No | ||
| 77 | PARVA | 730136 | 16466 | -0.940 | -0.5782 | No | ||
| 78 | FBXL17 | 770170 | 16485 | -0.953 | -0.5718 | No | ||
| 79 | PPP1R3F | 2450114 | 16810 | -1.175 | -0.5802 | Yes | ||
| 80 | SEC24C | 6980707 | 16811 | -1.177 | -0.5711 | Yes | ||
| 81 | RARA | 4050161 | 16922 | -1.268 | -0.5672 | Yes | ||
| 82 | TOMM34 | 5690102 | 16937 | -1.282 | -0.5581 | Yes | ||
| 83 | RGS16 | 780091 | 16984 | -1.327 | -0.5503 | Yes | ||
| 84 | LIMK2 | 2650446 4050164 | 17035 | -1.382 | -0.5424 | Yes | ||
| 85 | B4GALT3 | 6040064 | 17156 | -1.539 | -0.5370 | Yes | ||
| 86 | SMARCD2 | 3830619 | 17158 | -1.544 | -0.5251 | Yes | ||
| 87 | BCL2L1 | 1580452 4200152 5420484 | 17203 | -1.597 | -0.5151 | Yes | ||
| 88 | YPEL1 | 6590435 | 17232 | -1.635 | -0.5040 | Yes | ||
| 89 | PHF13 | 450435 | 17574 | -2.159 | -0.5058 | Yes | ||
| 90 | TXNL4B | 1690750 | 17747 | -2.518 | -0.4956 | Yes | ||
| 91 | SBK1 | 940452 | 17764 | -2.554 | -0.4767 | Yes | ||
| 92 | UHRF2 | 60050 840280 1340746 5080091 | 17842 | -2.723 | -0.4599 | Yes | ||
| 93 | JUNB | 4230048 | 17886 | -2.809 | -0.4405 | Yes | ||
| 94 | DNAJB5 | 2900215 | 17996 | -3.071 | -0.4226 | Yes | ||
| 95 | UBAP1 | 4280059 | 18020 | -3.122 | -0.3998 | Yes | ||
| 96 | GPRASP2 | 3990541 | 18058 | -3.294 | -0.3763 | Yes | ||
| 97 | ZFAND3 | 2760504 | 18142 | -3.617 | -0.3529 | Yes | ||
| 98 | RANBP10 | 2340184 | 18174 | -3.722 | -0.3258 | Yes | ||
| 99 | BRD2 | 1450300 5340484 6100605 | 18266 | -4.192 | -0.2983 | Yes | ||
| 100 | PPP1R9B | 3130619 | 18306 | -4.410 | -0.2664 | Yes | ||
| 101 | KLF13 | 4670563 | 18309 | -4.420 | -0.2323 | Yes | ||
| 102 | LPHN1 | 1400113 3130301 | 18320 | -4.479 | -0.1983 | Yes | ||
| 103 | XBP1 | 3840594 | 18340 | -4.607 | -0.1637 | Yes | ||
| 104 | MAP3K3 | 610685 | 18366 | -4.757 | -0.1283 | Yes | ||
| 105 | PLAGL2 | 1770148 | 18469 | -5.739 | -0.0895 | Yes | ||
| 106 | RNF44 | 5910692 | 18480 | -5.836 | -0.0449 | Yes | ||
| 107 | PHF2 | 4060300 | 18540 | -6.759 | 0.0041 | Yes |