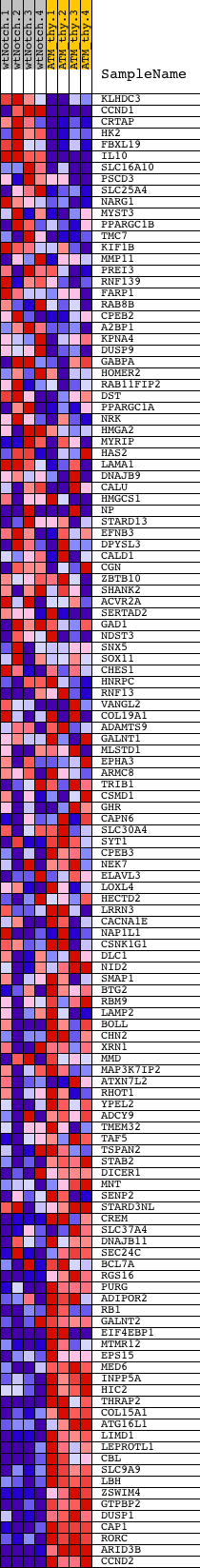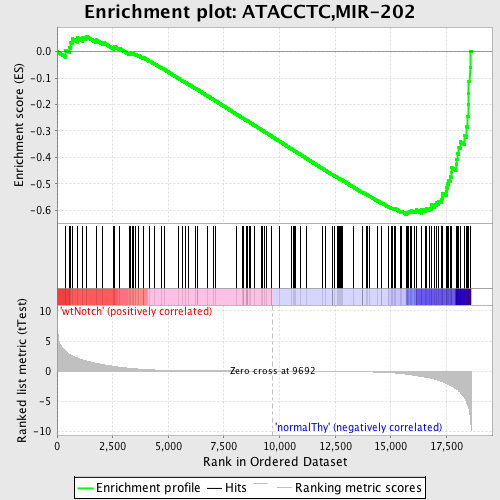
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy_repos |
| Phenotype | phenotype_wtNotch_versus_normalThy.cls#wtNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | ATACCTC,MIR-202 |
| Enrichment Score (ES) | -0.61527574 |
| Normalized Enrichment Score (NES) | -1.5956172 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.07778328 |
| FWER p-Value | 0.351 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | KLHDC3 | 3830717 | 386 | 3.274 | 0.0038 | No | ||
| 2 | CCND1 | 460524 770309 3120576 6980398 | 547 | 2.823 | 0.0164 | No | ||
| 3 | CRTAP | 3990725 | 619 | 2.666 | 0.0326 | No | ||
| 4 | HK2 | 2640722 | 673 | 2.572 | 0.0491 | No | ||
| 5 | FBXL19 | 4570039 | 921 | 2.153 | 0.0520 | No | ||
| 6 | IL10 | 2340685 2640541 2850403 6590286 | 1156 | 1.855 | 0.0533 | No | ||
| 7 | SLC16A10 | 4010672 | 1303 | 1.712 | 0.0583 | No | ||
| 8 | PSCD3 | 730301 | 1750 | 1.304 | 0.0440 | No | ||
| 9 | SLC25A4 | 2360519 | 2057 | 1.060 | 0.0354 | No | ||
| 10 | NARG1 | 5910563 6350095 | 2543 | 0.794 | 0.0151 | No | ||
| 11 | MYST3 | 5270500 | 2576 | 0.772 | 0.0192 | No | ||
| 12 | PPARGC1B | 3290114 | 2786 | 0.666 | 0.0129 | No | ||
| 13 | TMC7 | 1990129 5290253 | 3237 | 0.485 | -0.0078 | No | ||
| 14 | KIF1B | 1240494 2370139 4570270 6510102 | 3307 | 0.455 | -0.0081 | No | ||
| 15 | MMP11 | 1980619 | 3396 | 0.431 | -0.0096 | No | ||
| 16 | PREI3 | 3170066 3440075 6370427 | 3414 | 0.425 | -0.0073 | No | ||
| 17 | RNF139 | 2650114 3610450 | 3528 | 0.383 | -0.0105 | No | ||
| 18 | FARP1 | 5290102 7100341 | 3678 | 0.330 | -0.0161 | No | ||
| 19 | RAB8B | 510692 1980017 3130136 4570739 | 3865 | 0.283 | -0.0240 | No | ||
| 20 | CPEB2 | 4760338 | 3872 | 0.282 | -0.0222 | No | ||
| 21 | A2BP1 | 2370390 4590593 5550014 | 4173 | 0.223 | -0.0368 | No | ||
| 22 | KPNA4 | 510369 | 4355 | 0.198 | -0.0451 | No | ||
| 23 | DUSP9 | 630605 | 4680 | 0.161 | -0.0614 | No | ||
| 24 | GABPA | 2360017 4780301 4850279 | 4835 | 0.146 | -0.0686 | No | ||
| 25 | HOMER2 | 840242 | 5473 | 0.103 | -0.1023 | No | ||
| 26 | RAB11FIP2 | 4570040 | 5645 | 0.095 | -0.1108 | No | ||
| 27 | DST | 430026 1090035 2340577 3170068 3870112 4780519 6400167 6450358 7040347 | 5757 | 0.089 | -0.1162 | No | ||
| 28 | PPARGC1A | 4670040 | 5917 | 0.082 | -0.1242 | No | ||
| 29 | NRK | 4590609 | 6210 | 0.071 | -0.1394 | No | ||
| 30 | HMGA2 | 2940121 3390647 5130279 6400136 | 6308 | 0.067 | -0.1442 | No | ||
| 31 | MYRIP | 1580471 | 6330 | 0.067 | -0.1448 | No | ||
| 32 | HAS2 | 5360181 | 6744 | 0.054 | -0.1667 | No | ||
| 33 | LAMA1 | 1770594 | 7008 | 0.047 | -0.1806 | No | ||
| 34 | DNAJB9 | 460114 | 7132 | 0.044 | -0.1869 | No | ||
| 35 | CALU | 2680403 3520056 4610348 5720176 6130121 | 8083 | 0.026 | -0.2381 | No | ||
| 36 | HMGCS1 | 6620452 | 8346 | 0.021 | -0.2521 | No | ||
| 37 | NP | 2630577 3120142 5670068 6290039 | 8368 | 0.021 | -0.2531 | No | ||
| 38 | STARD13 | 540722 | 8392 | 0.021 | -0.2542 | No | ||
| 39 | EFNB3 | 5570594 | 8501 | 0.019 | -0.2599 | No | ||
| 40 | DPYSL3 | 1580594 | 8516 | 0.018 | -0.2605 | No | ||
| 41 | CALD1 | 1770129 1940397 | 8546 | 0.018 | -0.2619 | No | ||
| 42 | CGN | 5890139 6370167 6400537 | 8549 | 0.018 | -0.2619 | No | ||
| 43 | ZBTB10 | 4200132 | 8659 | 0.016 | -0.2677 | No | ||
| 44 | SHANK2 | 1500452 | 8684 | 0.016 | -0.2689 | No | ||
| 45 | ACVR2A | 6110647 | 8879 | 0.012 | -0.2793 | No | ||
| 46 | SERTAD2 | 2810673 6840450 | 9200 | 0.007 | -0.2965 | No | ||
| 47 | GAD1 | 2360035 3140167 | 9221 | 0.007 | -0.2976 | No | ||
| 48 | NDST3 | 670020 | 9319 | 0.006 | -0.3028 | No | ||
| 49 | SNX5 | 1570021 6860609 | 9426 | 0.004 | -0.3085 | No | ||
| 50 | SOX11 | 610279 | 9618 | 0.001 | -0.3188 | No | ||
| 51 | CHES1 | 4570121 5700162 | 9641 | 0.001 | -0.3200 | No | ||
| 52 | HNRPC | 1570133 3610131 | 9994 | -0.004 | -0.3390 | No | ||
| 53 | RNF13 | 2370021 | 10545 | -0.012 | -0.3686 | No | ||
| 54 | VANGL2 | 70097 870075 | 10610 | -0.014 | -0.3720 | No | ||
| 55 | COL19A1 | 6220441 | 10649 | -0.014 | -0.3740 | No | ||
| 56 | ADAMTS9 | 2760079 | 10731 | -0.015 | -0.3782 | No | ||
| 57 | GALNT1 | 4070487 6130100 6250132 6900048 | 10925 | -0.018 | -0.3885 | No | ||
| 58 | MLSTD1 | 3710184 | 11215 | -0.023 | -0.4040 | No | ||
| 59 | EPHA3 | 2190497 5080059 | 11906 | -0.036 | -0.4410 | No | ||
| 60 | ARMC8 | 520408 940132 4060368 4210102 | 12065 | -0.040 | -0.4493 | No | ||
| 61 | TRIB1 | 2320435 | 12355 | -0.047 | -0.4646 | No | ||
| 62 | CSMD1 | 1780672 | 12363 | -0.047 | -0.4646 | No | ||
| 63 | GHR | 2260156 | 12369 | -0.047 | -0.4645 | No | ||
| 64 | CAPN6 | 1740168 | 12474 | -0.049 | -0.4697 | No | ||
| 65 | SLC30A4 | 1850372 | 12582 | -0.053 | -0.4751 | No | ||
| 66 | SYT1 | 840364 | 12644 | -0.055 | -0.4780 | No | ||
| 67 | CPEB3 | 3940164 | 12711 | -0.056 | -0.4812 | No | ||
| 68 | NEK7 | 4540026 | 12729 | -0.057 | -0.4817 | No | ||
| 69 | ELAVL3 | 2850014 | 12776 | -0.059 | -0.4837 | No | ||
| 70 | LOXL4 | 6520095 | 12807 | -0.060 | -0.4849 | No | ||
| 71 | HECTD2 | 2680014 3060689 | 13309 | -0.079 | -0.5114 | No | ||
| 72 | LRRN3 | 1450133 | 13707 | -0.100 | -0.5321 | No | ||
| 73 | CACNA1E | 840253 | 13730 | -0.101 | -0.5325 | No | ||
| 74 | NAP1L1 | 2510593 5900215 5090470 450079 4920280 | 13732 | -0.101 | -0.5318 | No | ||
| 75 | CSNK1G1 | 840082 1230575 3940647 | 13886 | -0.112 | -0.5393 | No | ||
| 76 | DLC1 | 1090632 6450594 | 13941 | -0.116 | -0.5413 | No | ||
| 77 | NID2 | 2320059 | 14051 | -0.125 | -0.5463 | No | ||
| 78 | SMAP1 | 840056 2360523 | 14420 | -0.162 | -0.5649 | No | ||
| 79 | BTG2 | 2350411 | 14579 | -0.176 | -0.5722 | No | ||
| 80 | RBM9 | 2510092 6290088 | 14598 | -0.180 | -0.5718 | No | ||
| 81 | LAMP2 | 1230402 1980373 | 14876 | -0.229 | -0.5850 | No | ||
| 82 | BOLL | 6100743 | 15049 | -0.266 | -0.5923 | No | ||
| 83 | CHN2 | 870528 | 15086 | -0.277 | -0.5922 | No | ||
| 84 | XRN1 | 5900093 | 15154 | -0.292 | -0.5936 | No | ||
| 85 | MMD | 3850239 | 15200 | -0.307 | -0.5938 | No | ||
| 86 | MAP3K7IP2 | 2340242 | 15454 | -0.386 | -0.6045 | No | ||
| 87 | ATXN7L2 | 4010161 | 15476 | -0.398 | -0.6027 | No | ||
| 88 | RHOT1 | 6760692 | 15710 | -0.501 | -0.6115 | Yes | ||
| 89 | YPEL2 | 5560609 | 15730 | -0.512 | -0.6087 | Yes | ||
| 90 | ADCY9 | 5690168 | 15767 | -0.529 | -0.6066 | Yes | ||
| 91 | TMEM32 | 1170452 | 15815 | -0.547 | -0.6051 | Yes | ||
| 92 | TAF5 | 3450288 5890193 6860435 | 15861 | -0.573 | -0.6032 | Yes | ||
| 93 | TSPAN2 | 3940161 | 15919 | -0.613 | -0.6016 | Yes | ||
| 94 | STAB2 | 4810452 | 16058 | -0.685 | -0.6039 | Yes | ||
| 95 | DICER1 | 540286 | 16132 | -0.731 | -0.6024 | Yes | ||
| 96 | MNT | 4920575 | 16146 | -0.736 | -0.5976 | Yes | ||
| 97 | SENP2 | 4540452 | 16384 | -0.889 | -0.6037 | Yes | ||
| 98 | STARD3NL | 7000114 | 16392 | -0.894 | -0.5973 | Yes | ||
| 99 | CREM | 840156 6380438 6660041 6660168 | 16542 | -0.993 | -0.5979 | Yes | ||
| 100 | SLC37A4 | 6400079 | 16591 | -1.010 | -0.5929 | Yes | ||
| 101 | DNAJB11 | 4150168 | 16758 | -1.133 | -0.5934 | Yes | ||
| 102 | SEC24C | 6980707 | 16811 | -1.177 | -0.5873 | Yes | ||
| 103 | BCL7A | 4560095 | 16824 | -1.188 | -0.5790 | Yes | ||
| 104 | RGS16 | 780091 | 16984 | -1.327 | -0.5776 | Yes | ||
| 105 | PURG | 1190593 | 17045 | -1.408 | -0.5703 | Yes | ||
| 106 | ADIPOR2 | 1510035 | 17152 | -1.536 | -0.5644 | Yes | ||
| 107 | RB1 | 5900338 | 17268 | -1.683 | -0.5580 | Yes | ||
| 108 | GALNT2 | 2260279 4060239 | 17308 | -1.728 | -0.5471 | Yes | ||
| 109 | EIF4EBP1 | 60132 5720148 | 17318 | -1.734 | -0.5345 | Yes | ||
| 110 | MTMR12 | 7000181 | 17502 | -2.035 | -0.5291 | Yes | ||
| 111 | EPS15 | 4200215 | 17516 | -2.059 | -0.5143 | Yes | ||
| 112 | MED6 | 6840500 | 17545 | -2.110 | -0.4999 | Yes | ||
| 113 | INPP5A | 5910195 | 17605 | -2.216 | -0.4864 | Yes | ||
| 114 | HIC2 | 4010433 | 17664 | -2.356 | -0.4718 | Yes | ||
| 115 | THRAP2 | 4920600 | 17710 | -2.437 | -0.4559 | Yes | ||
| 116 | COL15A1 | 6840113 | 17715 | -2.446 | -0.4377 | Yes | ||
| 117 | ATG16L1 | 5390292 | 17931 | -2.941 | -0.4272 | Yes | ||
| 118 | LIMD1 | 2480142 | 17953 | -2.980 | -0.4059 | Yes | ||
| 119 | LEPROTL1 | 6900110 | 18000 | -3.077 | -0.3852 | Yes | ||
| 120 | CBL | 6380068 | 18061 | -3.305 | -0.3636 | Yes | ||
| 121 | SLC9A9 | 1940309 | 18116 | -3.496 | -0.3402 | Yes | ||
| 122 | LBH | 70093 | 18305 | -4.390 | -0.3173 | Yes | ||
| 123 | ZSWIM4 | 6940195 | 18415 | -5.184 | -0.2841 | Yes | ||
| 124 | GTPBP2 | 2760750 | 18462 | -5.667 | -0.2440 | Yes | ||
| 125 | DUSP1 | 6860121 | 18476 | -5.793 | -0.2011 | Yes | ||
| 126 | CAP1 | 2650278 | 18491 | -5.943 | -0.1571 | Yes | ||
| 127 | RORC | 1740121 | 18500 | -6.120 | -0.1114 | Yes | ||
| 128 | ARID3B | 6650008 | 18558 | -7.239 | -0.0600 | Yes | ||
| 129 | CCND2 | 5340167 | 18589 | -8.378 | 0.0015 | Yes |