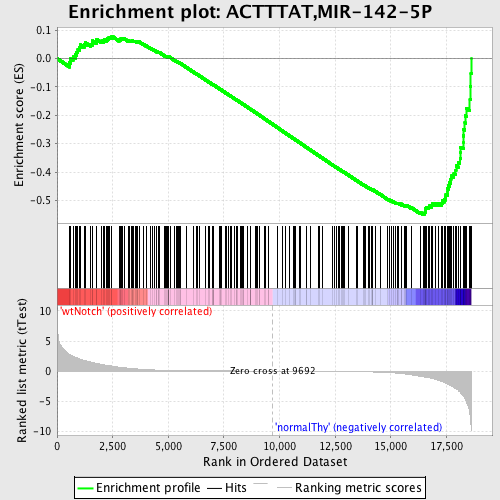
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy_repos |
| Phenotype | phenotype_wtNotch_versus_normalThy.cls#wtNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | ACTTTAT,MIR-142-5P |
| Enrichment Score (ES) | -0.5491195 |
| Normalized Enrichment Score (NES) | -1.5065988 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.12340283 |
| FWER p-Value | 0.815 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CELSR1 | 6510348 | 557 | 2.791 | -0.0147 | No | ||
| 2 | GCH1 | 670364 6550358 | 591 | 2.707 | -0.0014 | No | ||
| 3 | APRIN | 2810050 | 717 | 2.508 | 0.0057 | No | ||
| 4 | EGLN3 | 1660484 | 816 | 2.329 | 0.0134 | No | ||
| 5 | CAPN7 | 5550050 | 875 | 2.225 | 0.0226 | No | ||
| 6 | PPM1G | 610725 | 935 | 2.122 | 0.0312 | No | ||
| 7 | RNH1 | 840438 | 1007 | 2.021 | 0.0386 | No | ||
| 8 | HOXB4 | 540131 | 1039 | 1.987 | 0.0480 | No | ||
| 9 | DNAJC7 | 4920048 | 1214 | 1.797 | 0.0485 | No | ||
| 10 | RGMA | 2450711 | 1267 | 1.740 | 0.0554 | No | ||
| 11 | RAB11FIP5 | 4570600 | 1515 | 1.516 | 0.0504 | No | ||
| 12 | BTBD1 | 5720619 | 1579 | 1.453 | 0.0551 | No | ||
| 13 | HERPUD1 | 7100440 | 1584 | 1.449 | 0.0629 | No | ||
| 14 | BVES | 5340072 | 1771 | 1.292 | 0.0600 | No | ||
| 15 | MBIP | 2640524 5570441 | 1780 | 1.285 | 0.0667 | No | ||
| 16 | VPS54 | 1660168 | 1988 | 1.115 | 0.0617 | No | ||
| 17 | TGFBR2 | 1780711 1980537 6550398 | 2076 | 1.050 | 0.0628 | No | ||
| 18 | HSPA8 | 2120315 1050132 6550746 | 2111 | 1.030 | 0.0667 | No | ||
| 19 | OTX2 | 1190471 | 2239 | 0.983 | 0.0652 | No | ||
| 20 | TRIP10 | 460438 | 2265 | 0.971 | 0.0693 | No | ||
| 21 | ALS2 | 3130546 6400070 | 2307 | 0.941 | 0.0723 | No | ||
| 22 | TRIM36 | 1400600 1980082 5130037 | 2364 | 0.904 | 0.0743 | No | ||
| 23 | PBX3 | 1300424 3710577 6180575 | 2438 | 0.857 | 0.0751 | No | ||
| 24 | ACIN1 | 3390278 6350377 6620142 | 2459 | 0.845 | 0.0787 | No | ||
| 25 | SCOC | 610048 2230053 | 2784 | 0.667 | 0.0648 | No | ||
| 26 | GRSF1 | 2100184 | 2843 | 0.641 | 0.0652 | No | ||
| 27 | ZFX | 5900400 | 2845 | 0.641 | 0.0688 | No | ||
| 28 | CRSP2 | 1940059 | 2885 | 0.625 | 0.0701 | No | ||
| 29 | FBXL3 | 5910142 | 2926 | 0.604 | 0.0713 | No | ||
| 30 | PAIP1 | 3870576 | 3019 | 0.565 | 0.0695 | No | ||
| 31 | GAS7 | 2120358 6450494 | 3203 | 0.498 | 0.0623 | No | ||
| 32 | CAMK2A | 1740333 1940112 4150292 | 3263 | 0.474 | 0.0617 | No | ||
| 33 | SLC36A1 | 5080242 | 3327 | 0.450 | 0.0608 | No | ||
| 34 | NFAT5 | 2510411 5890195 6550152 | 3369 | 0.441 | 0.0610 | No | ||
| 35 | MEA1 | 2230347 6350338 | 3374 | 0.440 | 0.0633 | No | ||
| 36 | MGAT4B | 130053 6840100 | 3453 | 0.407 | 0.0613 | No | ||
| 37 | EGR3 | 6940128 | 3503 | 0.392 | 0.0608 | No | ||
| 38 | REV3L | 1090717 | 3576 | 0.367 | 0.0589 | No | ||
| 39 | SRI | 3390446 4850064 | 3614 | 0.351 | 0.0589 | No | ||
| 40 | CUL4A | 1170088 2320008 2470278 | 3616 | 0.349 | 0.0608 | No | ||
| 41 | STC1 | 360161 | 3684 | 0.328 | 0.0590 | No | ||
| 42 | CPEB2 | 4760338 | 3872 | 0.282 | 0.0504 | No | ||
| 43 | SNX16 | 870446 | 3996 | 0.253 | 0.0451 | No | ||
| 44 | MSH6 | 4480064 6520093 | 4194 | 0.219 | 0.0356 | No | ||
| 45 | ST8SIA2 | 2680082 | 4267 | 0.208 | 0.0329 | No | ||
| 46 | CD69 | 380167 4730088 | 4356 | 0.198 | 0.0292 | No | ||
| 47 | RHOC | 2060242 | 4478 | 0.181 | 0.0236 | No | ||
| 48 | NPAT | 3800594 4670026 | 4488 | 0.181 | 0.0241 | No | ||
| 49 | MMP24 | 5340154 | 4542 | 0.175 | 0.0222 | No | ||
| 50 | UCHL3 | 1240735 | 4574 | 0.171 | 0.0215 | No | ||
| 51 | CNNM4 | 1740347 | 4604 | 0.168 | 0.0209 | No | ||
| 52 | ARID4B | 730754 6660026 | 4844 | 0.145 | 0.0087 | No | ||
| 53 | IGF1 | 1990193 3130377 3290280 | 4888 | 0.141 | 0.0071 | No | ||
| 54 | STAG1 | 1190300 1400722 4590100 | 4925 | 0.138 | 0.0059 | No | ||
| 55 | SON | 3120148 5860239 7040403 | 4954 | 0.136 | 0.0052 | No | ||
| 56 | ABCG4 | 3440458 | 4991 | 0.132 | 0.0040 | No | ||
| 57 | FXYD3 | 2320450 | 4996 | 0.132 | 0.0045 | No | ||
| 58 | SLC18A2 | 6420541 | 5003 | 0.131 | 0.0049 | No | ||
| 59 | CBX3 | 1780592 3940008 | 5008 | 0.131 | 0.0054 | No | ||
| 60 | UBE4A | 6100520 | 5025 | 0.130 | 0.0052 | No | ||
| 61 | QKI | 5220093 6130044 | 5026 | 0.130 | 0.0060 | No | ||
| 62 | RPS6KA4 | 2190524 | 5105 | 0.125 | 0.0024 | No | ||
| 63 | UBR1 | 3800132 | 5279 | 0.114 | -0.0063 | No | ||
| 64 | TRIM3 | 5570156 | 5367 | 0.109 | -0.0105 | No | ||
| 65 | LRP1B | 1450193 | 5428 | 0.106 | -0.0131 | No | ||
| 66 | SLC4A4 | 1780075 1850040 4810068 | 5438 | 0.105 | -0.0130 | No | ||
| 67 | HMGCLL1 | 2570091 | 5514 | 0.101 | -0.0166 | No | ||
| 68 | COL4A3 | 5910075 | 5555 | 0.099 | -0.0182 | No | ||
| 69 | SLC25A27 | 6200707 6940019 | 5566 | 0.099 | -0.0182 | No | ||
| 70 | FNDC3A | 2690097 3290044 | 5805 | 0.086 | -0.0306 | No | ||
| 71 | CCNT2 | 2100390 4150563 | 6114 | 0.074 | -0.0469 | No | ||
| 72 | MKLN1 | 70504 1850129 | 6242 | 0.069 | -0.0534 | No | ||
| 73 | CUL2 | 4200278 | 6288 | 0.068 | -0.0555 | No | ||
| 74 | ADAMTS5 | 5890592 | 6380 | 0.065 | -0.0601 | No | ||
| 75 | STK35 | 2360121 | 6675 | 0.056 | -0.0758 | No | ||
| 76 | HIAT1 | 1580176 | 6820 | 0.052 | -0.0833 | No | ||
| 77 | PAPPA | 4230463 | 6839 | 0.051 | -0.0840 | No | ||
| 78 | RERG | 5910324 | 6974 | 0.048 | -0.0910 | No | ||
| 79 | KLF10 | 4850056 | 6976 | 0.048 | -0.0908 | No | ||
| 80 | PURA | 3870156 | 7018 | 0.047 | -0.0927 | No | ||
| 81 | SREBF1 | 4780333 | 7022 | 0.047 | -0.0926 | No | ||
| 82 | EXOC5 | 7000711 | 7034 | 0.047 | -0.0930 | No | ||
| 83 | SIX4 | 6760022 | 7278 | 0.041 | -0.1060 | No | ||
| 84 | RTN1 | 4150452 4850176 | 7324 | 0.040 | -0.1082 | No | ||
| 85 | ETV1 | 70735 2940603 5080463 | 7339 | 0.040 | -0.1087 | No | ||
| 86 | AP1G1 | 2360671 | 7399 | 0.039 | -0.1117 | No | ||
| 87 | KLF11 | 6840739 | 7586 | 0.035 | -0.1216 | No | ||
| 88 | ZCCHC11 | 3390731 5360039 | 7627 | 0.034 | -0.1236 | No | ||
| 89 | NPAS4 | 2320204 4150154 | 7717 | 0.033 | -0.1282 | No | ||
| 90 | RNF128 | 1230300 5670273 | 7805 | 0.031 | -0.1328 | No | ||
| 91 | GOPC | 6620400 | 7838 | 0.031 | -0.1344 | No | ||
| 92 | HDLBP | 2350692 6590520 | 7982 | 0.028 | -0.1420 | No | ||
| 93 | KCNJ3 | 50184 | 8043 | 0.027 | -0.1451 | No | ||
| 94 | BMP2 | 6620687 | 8045 | 0.027 | -0.1450 | No | ||
| 95 | DIO2 | 380504 540403 | 8109 | 0.025 | -0.1483 | No | ||
| 96 | EPAS1 | 5290156 | 8237 | 0.023 | -0.1550 | No | ||
| 97 | FZD7 | 7050706 | 8277 | 0.022 | -0.1570 | No | ||
| 98 | MASTL | 380433 4480215 5390301 | 8338 | 0.022 | -0.1602 | No | ||
| 99 | LRP2 | 1400204 3290706 | 8362 | 0.021 | -0.1613 | No | ||
| 100 | SLC17A7 | 7000037 | 8395 | 0.020 | -0.1629 | No | ||
| 101 | LHX8 | 5700347 | 8571 | 0.017 | -0.1723 | No | ||
| 102 | SHANK2 | 1500452 | 8684 | 0.016 | -0.1783 | No | ||
| 103 | BTG3 | 7050079 | 8689 | 0.016 | -0.1785 | No | ||
| 104 | GPR88 | 2450435 | 8690 | 0.016 | -0.1784 | No | ||
| 105 | BHLHB3 | 3450438 | 8696 | 0.015 | -0.1786 | No | ||
| 106 | RCN2 | 840324 | 8895 | 0.012 | -0.1893 | No | ||
| 107 | ALCAM | 1050019 | 8961 | 0.011 | -0.1927 | No | ||
| 108 | ZFPM2 | 460072 | 9001 | 0.010 | -0.1948 | No | ||
| 109 | MBD2 | 5900546 | 9085 | 0.009 | -0.1993 | No | ||
| 110 | HOXA13 | 3120647 | 9108 | 0.009 | -0.2004 | No | ||
| 111 | LUZP2 | 60673 | 9311 | 0.006 | -0.2113 | No | ||
| 112 | PAPOLB | 3840450 | 9371 | 0.005 | -0.2145 | No | ||
| 113 | BAI3 | 940692 | 9500 | 0.003 | -0.2215 | No | ||
| 114 | SPOCK2 | 6650369 | 9521 | 0.003 | -0.2225 | No | ||
| 115 | UNC84A | 1450632 3520408 | 9923 | -0.003 | -0.2443 | No | ||
| 116 | RHOQ | 520161 | 10151 | -0.007 | -0.2566 | No | ||
| 117 | BTBD7 | 730288 | 10256 | -0.008 | -0.2622 | No | ||
| 118 | PTPN4 | 1410047 | 10433 | -0.011 | -0.2717 | No | ||
| 119 | UBE2D1 | 1850400 | 10642 | -0.014 | -0.2829 | No | ||
| 120 | NRG1 | 1050332 | 10681 | -0.015 | -0.2849 | No | ||
| 121 | TAOK2 | 110575 6380692 | 10719 | -0.015 | -0.2868 | No | ||
| 122 | EYA4 | 5360286 | 10726 | -0.015 | -0.2871 | No | ||
| 123 | ABCA1 | 6290156 | 10877 | -0.018 | -0.2951 | No | ||
| 124 | ARID2 | 110647 5390435 | 10958 | -0.019 | -0.2994 | No | ||
| 125 | FIGN | 7000601 | 11187 | -0.023 | -0.3116 | No | ||
| 126 | NTRK3 | 4050687 4070167 4610026 6100484 | 11202 | -0.023 | -0.3122 | No | ||
| 127 | SYT7 | 2450528 3390500 3610687 | 11374 | -0.026 | -0.3214 | No | ||
| 128 | DCHS1 | 4850487 | 11762 | -0.034 | -0.3422 | No | ||
| 129 | ZCCHC14 | 5900288 | 11806 | -0.034 | -0.3444 | No | ||
| 130 | ELAVL2 | 360181 | 11943 | -0.037 | -0.3515 | No | ||
| 131 | JMJD1C | 940575 2120025 | 12368 | -0.047 | -0.3743 | No | ||
| 132 | ADAMTS1 | 3140286 5860463 | 12472 | -0.049 | -0.3796 | No | ||
| 133 | PRPF4B | 940181 2570300 | 12561 | -0.052 | -0.3841 | No | ||
| 134 | MYF5 | 1050739 | 12664 | -0.055 | -0.3894 | No | ||
| 135 | CPEB3 | 3940164 | 12711 | -0.056 | -0.3916 | No | ||
| 136 | MYO1D | 6760292 | 12779 | -0.059 | -0.3949 | No | ||
| 137 | RIPK5 | 1230647 1340114 | 12821 | -0.060 | -0.3968 | No | ||
| 138 | ROBO1 | 3710398 | 12871 | -0.062 | -0.3991 | No | ||
| 139 | GARNL1 | 5670398 | 12895 | -0.062 | -0.4000 | No | ||
| 140 | TPBG | 630161 | 13086 | -0.069 | -0.4099 | No | ||
| 141 | VANGL1 | 380162 | 13453 | -0.086 | -0.4293 | No | ||
| 142 | KITLG | 2120047 6220300 | 13497 | -0.088 | -0.4312 | No | ||
| 143 | GRM7 | 4230110 | 13782 | -0.105 | -0.4460 | No | ||
| 144 | WDR26 | 1090138 | 13808 | -0.107 | -0.4468 | No | ||
| 145 | AHR | 4560671 | 13844 | -0.110 | -0.4481 | No | ||
| 146 | KCNRG | 3460315 3940072 | 13975 | -0.119 | -0.4545 | No | ||
| 147 | LMX1A | 2810239 5690324 | 14024 | -0.122 | -0.4564 | No | ||
| 148 | RPS6KA5 | 2120563 7040546 | 14110 | -0.130 | -0.4603 | No | ||
| 149 | MAT2B | 1690139 2510706 | 14187 | -0.137 | -0.4636 | No | ||
| 150 | DDHD1 | 3450241 3940523 6380156 | 14189 | -0.137 | -0.4629 | No | ||
| 151 | RAB6B | 5860121 | 14194 | -0.138 | -0.4624 | No | ||
| 152 | SOX5 | 2370576 2900167 3190128 5050528 | 14330 | -0.152 | -0.4689 | No | ||
| 153 | CBLN4 | 4010451 | 14531 | -0.171 | -0.4788 | No | ||
| 154 | PLEKHA3 | 5080068 | 14863 | -0.226 | -0.4955 | No | ||
| 155 | SP2 | 6400026 | 14931 | -0.243 | -0.4978 | No | ||
| 156 | FOS | 1850315 | 15011 | -0.260 | -0.5006 | No | ||
| 157 | PCTK2 | 5700673 | 15099 | -0.279 | -0.5038 | No | ||
| 158 | PCBP2 | 5690048 | 15222 | -0.316 | -0.5087 | No | ||
| 159 | CHD9 | 430037 2570129 5420398 | 15277 | -0.333 | -0.5098 | No | ||
| 160 | PLXNA2 | 6450132 | 15321 | -0.344 | -0.5102 | No | ||
| 161 | TCF12 | 3610324 7000156 | 15344 | -0.352 | -0.5094 | No | ||
| 162 | MGLL | 2030446 4210520 | 15472 | -0.398 | -0.5141 | No | ||
| 163 | ATXN7L2 | 4010161 | 15476 | -0.398 | -0.5120 | No | ||
| 164 | RPS25 | 520139 | 15616 | -0.457 | -0.5171 | No | ||
| 165 | NFE2L2 | 2810619 3390162 | 15653 | -0.473 | -0.5164 | No | ||
| 166 | RHOT1 | 6760692 | 15710 | -0.501 | -0.5166 | No | ||
| 167 | ELAVL4 | 50735 3360086 5220167 | 15928 | -0.615 | -0.5250 | No | ||
| 168 | KIT | 7040095 | 16331 | -0.857 | -0.5421 | No | ||
| 169 | IL18BP | 3450128 | 16462 | -0.938 | -0.5439 | Yes | ||
| 170 | CRK | 1230162 4780128 | 16534 | -0.989 | -0.5423 | Yes | ||
| 171 | SDCBP | 460487 2190039 5270441 | 16559 | -0.999 | -0.5380 | Yes | ||
| 172 | BNIP2 | 1410475 6770088 | 16563 | -0.999 | -0.5326 | Yes | ||
| 173 | CLCN3 | 510195 780176 1660093 3830372 | 16568 | -1.001 | -0.5272 | Yes | ||
| 174 | SRF | 70139 2320039 | 16612 | -1.017 | -0.5239 | Yes | ||
| 175 | PCBP1 | 1050088 | 16705 | -1.090 | -0.5228 | Yes | ||
| 176 | HIPK1 | 110193 | 16748 | -1.122 | -0.5189 | Yes | ||
| 177 | H3F3A | 2900086 | 16833 | -1.191 | -0.5168 | Yes | ||
| 178 | APBB3 | 6020168 | 16863 | -1.214 | -0.5116 | Yes | ||
| 179 | OSR1 | 1500025 | 17000 | -1.345 | -0.5115 | Yes | ||
| 180 | SSH2 | 360056 | 17136 | -1.523 | -0.5104 | Yes | ||
| 181 | FGF13 | 630575 1570440 5360121 | 17286 | -1.707 | -0.5090 | Yes | ||
| 182 | RHOA | 580142 5900131 5340450 | 17313 | -1.730 | -0.5008 | Yes | ||
| 183 | MED28 | 3420025 | 17398 | -1.868 | -0.4949 | Yes | ||
| 184 | UBE2A | 4010563 | 17462 | -1.967 | -0.4874 | Yes | ||
| 185 | BAZ2A | 730184 | 17477 | -1.996 | -0.4771 | Yes | ||
| 186 | BECN1 | 3840433 | 17553 | -2.125 | -0.4693 | Yes | ||
| 187 | DUSP2 | 2370184 | 17558 | -2.134 | -0.4576 | Yes | ||
| 188 | HN1 | 3360156 | 17603 | -2.212 | -0.4477 | Yes | ||
| 189 | CDK5 | 940348 | 17639 | -2.315 | -0.4367 | Yes | ||
| 190 | LRP12 | 2370112 | 17690 | -2.398 | -0.4261 | Yes | ||
| 191 | TAF10 | 2760020 | 17706 | -2.435 | -0.4134 | Yes | ||
| 192 | CCNG2 | 3190095 | 17836 | -2.706 | -0.4053 | Yes | ||
| 193 | ULK1 | 6100315 | 17921 | -2.898 | -0.3937 | Yes | ||
| 194 | ATG16L1 | 5390292 | 17931 | -2.941 | -0.3779 | Yes | ||
| 195 | FOXJ2 | 2450441 | 18029 | -3.166 | -0.3655 | Yes | ||
| 196 | ACTN4 | 3840301 4590390 7050132 | 18109 | -3.462 | -0.3505 | Yes | ||
| 197 | TLE4 | 1450064 1500519 | 18134 | -3.567 | -0.3320 | Yes | ||
| 198 | ZFAND3 | 2760504 | 18142 | -3.617 | -0.3122 | Yes | ||
| 199 | CIC | 6370161 | 18256 | -4.145 | -0.2953 | Yes | ||
| 200 | SPEN | 2060041 | 18262 | -4.181 | -0.2723 | Yes | ||
| 201 | ATP1B1 | 3130594 | 18279 | -4.259 | -0.2495 | Yes | ||
| 202 | MCL1 | 1660672 | 18331 | -4.573 | -0.2268 | Yes | ||
| 203 | UBE3C | 3140673 | 18354 | -4.694 | -0.2019 | Yes | ||
| 204 | ZFYVE21 | 1940056 | 18405 | -5.095 | -0.1762 | Yes | ||
| 205 | LRRIQ2 | 3990064 6370673 6380372 | 18557 | -7.213 | -0.1443 | Yes | ||
| 206 | TRPC4AP | 840520 1660402 | 18588 | -8.365 | -0.0993 | Yes | ||
| 207 | ZFP36 | 2030605 | 18601 | -8.743 | -0.0513 | Yes | ||
| 208 | BTG1 | 4200735 6040131 6200133 | 18614 | -9.359 | 0.0001 | Yes |

