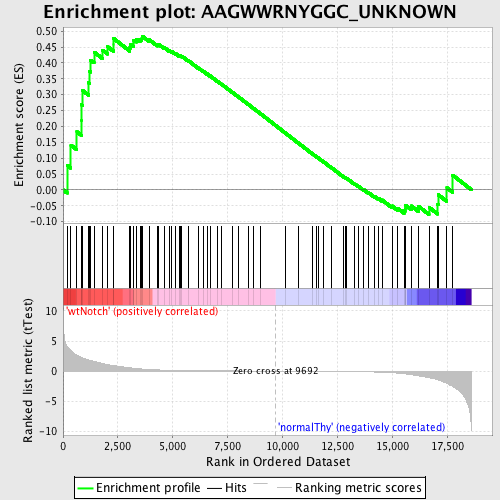
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy_repos |
| Phenotype | phenotype_wtNotch_versus_normalThy.cls#wtNotch_versus_normalThy_repos |
| Upregulated in class | wtNotch |
| GeneSet | AAGWWRNYGGC_UNKNOWN |
| Enrichment Score (ES) | 0.48471448 |
| Normalized Enrichment Score (NES) | 1.2284474 |
| Nominal p-value | 0.10093897 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MRPL38 | 1940154 | 184 | 4.129 | 0.0780 | Yes | ||
| 2 | SUPT16H | 1240333 | 357 | 3.397 | 0.1411 | Yes | ||
| 3 | SMYD5 | 6220025 | 629 | 2.654 | 0.1830 | Yes | ||
| 4 | RFC1 | 1190673 | 840 | 2.283 | 0.2203 | Yes | ||
| 5 | PSMD8 | 630142 | 846 | 2.266 | 0.2682 | Yes | ||
| 6 | CDC42BPB | 6350037 | 884 | 2.210 | 0.3133 | Yes | ||
| 7 | CRSP8 | 1450546 | 1159 | 1.848 | 0.3379 | Yes | ||
| 8 | DNAJC7 | 4920048 | 1214 | 1.797 | 0.3733 | Yes | ||
| 9 | ARL1 | 450619 | 1251 | 1.757 | 0.4088 | Yes | ||
| 10 | RHOB | 1500309 | 1412 | 1.615 | 0.4345 | Yes | ||
| 11 | NASP | 2260139 2940369 5130707 | 1798 | 1.270 | 0.4408 | Yes | ||
| 12 | CDKN2C | 5050750 5130148 | 2033 | 1.076 | 0.4511 | Yes | ||
| 13 | POLK | 5890270 | 2293 | 0.950 | 0.4573 | Yes | ||
| 14 | UBE2D3 | 3190452 | 2294 | 0.950 | 0.4776 | Yes | ||
| 15 | POLR2A | 5220647 | 3043 | 0.556 | 0.4491 | Yes | ||
| 16 | RALA | 2680471 | 3084 | 0.544 | 0.4585 | Yes | ||
| 17 | RAB22A | 110500 3830707 | 3217 | 0.492 | 0.4619 | Yes | ||
| 18 | IRAK1 | 4120593 | 3218 | 0.492 | 0.4723 | Yes | ||
| 19 | ARCN1 | 5820273 | 3336 | 0.448 | 0.4756 | Yes | ||
| 20 | CXXC1 | 630170 3940136 | 3511 | 0.389 | 0.4745 | Yes | ||
| 21 | TSC1 | 1850672 | 3593 | 0.359 | 0.4778 | Yes | ||
| 22 | UBE3A | 1240152 2690438 5860609 | 3605 | 0.355 | 0.4847 | Yes | ||
| 23 | CNOT3 | 130519 4480292 1990609 | 3924 | 0.271 | 0.4733 | No | ||
| 24 | ATF1 | 4570142 5550095 | 4294 | 0.206 | 0.4578 | No | ||
| 25 | COL4A3BP | 3060176 4670541 | 4346 | 0.199 | 0.4593 | No | ||
| 26 | HOXA9 | 610494 4730040 5130601 | 4619 | 0.166 | 0.4482 | No | ||
| 27 | CLSTN1 | 6020019 | 4853 | 0.145 | 0.4387 | No | ||
| 28 | ARF1 | 3840291 | 4955 | 0.135 | 0.4361 | No | ||
| 29 | CHD4 | 5420059 6130338 6380717 | 5129 | 0.123 | 0.4294 | No | ||
| 30 | NEUROD1 | 3060619 | 5284 | 0.114 | 0.4236 | No | ||
| 31 | OTX1 | 2510215 | 5342 | 0.111 | 0.4228 | No | ||
| 32 | HOXB6 | 4210739 | 5417 | 0.106 | 0.4211 | No | ||
| 33 | SLITRK3 | 5910041 | 5708 | 0.092 | 0.4074 | No | ||
| 34 | RNF12 | 6860717 | 6163 | 0.072 | 0.3845 | No | ||
| 35 | CHCHD7 | 6620692 | 6385 | 0.064 | 0.3739 | No | ||
| 36 | HOXC10 | 5360170 | 6559 | 0.059 | 0.3659 | No | ||
| 37 | PHF8 | 70722 | 6698 | 0.055 | 0.3596 | No | ||
| 38 | ZBTB4 | 6450441 | 7042 | 0.046 | 0.3421 | No | ||
| 39 | TIA1 | 4670373 | 7212 | 0.042 | 0.3339 | No | ||
| 40 | RAD23A | 670082 5270309 5360463 5550364 | 7712 | 0.033 | 0.3077 | No | ||
| 41 | RNF26 | 3710292 | 7976 | 0.028 | 0.2941 | No | ||
| 42 | NRAS | 6900577 | 8445 | 0.019 | 0.2693 | No | ||
| 43 | SHANK2 | 1500452 | 8684 | 0.016 | 0.2568 | No | ||
| 44 | PIGN | 4210736 | 9005 | 0.010 | 0.2397 | No | ||
| 45 | SUV39H2 | 1450672 6370242 | 10132 | -0.006 | 0.1791 | No | ||
| 46 | ADAMTS9 | 2760079 | 10731 | -0.015 | 0.1472 | No | ||
| 47 | HOXA2 | 2120121 | 11363 | -0.026 | 0.1137 | No | ||
| 48 | PAX6 | 1190025 | 11552 | -0.029 | 0.1042 | No | ||
| 49 | NFYC | 1400746 | 11627 | -0.031 | 0.1009 | No | ||
| 50 | STRN4 | 3440711 | 11871 | -0.036 | 0.0885 | No | ||
| 51 | BNC2 | 4810603 | 12219 | -0.044 | 0.0707 | No | ||
| 52 | PRDM1 | 3170347 3520301 | 12786 | -0.059 | 0.0415 | No | ||
| 53 | APBA1 | 3140487 | 12856 | -0.061 | 0.0391 | No | ||
| 54 | PLAG1 | 1450142 3870139 6520039 | 12927 | -0.064 | 0.0366 | No | ||
| 55 | YY1 | 6130451 | 13298 | -0.079 | 0.0184 | No | ||
| 56 | EIF5 | 6380750 | 13457 | -0.086 | 0.0117 | No | ||
| 57 | LRRN3 | 1450133 | 13707 | -0.100 | 0.0004 | No | ||
| 58 | RFX3 | 520121 4280519 5570519 6100184 | 13915 | -0.114 | -0.0084 | No | ||
| 59 | CNOT4 | 2320242 6760706 | 14209 | -0.140 | -0.0212 | No | ||
| 60 | MAFA | 70519 | 14391 | -0.157 | -0.0276 | No | ||
| 61 | RBM3 | 2360739 5700167 | 14537 | -0.172 | -0.0318 | No | ||
| 62 | NKIRAS2 | 580519 | 14989 | -0.256 | -0.0506 | No | ||
| 63 | BCL2L2 | 2760692 6770739 | 15246 | -0.324 | -0.0575 | No | ||
| 64 | MYLIP | 50717 4050672 | 15540 | -0.428 | -0.0642 | No | ||
| 65 | VAMP2 | 2100100 | 15586 | -0.445 | -0.0572 | No | ||
| 66 | SFN | 6290301 7510608 | 15592 | -0.446 | -0.0479 | No | ||
| 67 | FMO5 | 4810725 5390086 | 15872 | -0.584 | -0.0506 | No | ||
| 68 | PPARBP | 4280131 5690022 5860050 | 16200 | -0.771 | -0.0518 | No | ||
| 69 | EPC1 | 1570050 5290095 6900193 | 16678 | -1.065 | -0.0548 | No | ||
| 70 | RPS27 | 5270242 | 17075 | -1.446 | -0.0454 | No | ||
| 71 | SFPQ | 4760110 | 17096 | -1.466 | -0.0152 | No | ||
| 72 | FKRP | 130546 | 17494 | -2.021 | 0.0064 | No | ||
| 73 | TRIM8 | 6130441 | 17757 | -2.538 | 0.0463 | No |

