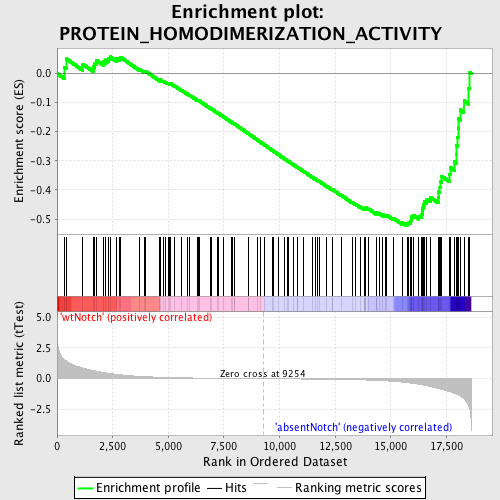
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #wtNotch_versus_absentNotch |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#wtNotch_versus_absentNotch |
| Upregulated in class | absentNotch |
| GeneSet | PROTEIN_HOMODIMERIZATION_ACTIVITY |
| Enrichment Score (ES) | -0.523409 |
| Normalized Enrichment Score (NES) | -1.504482 |
| Nominal p-value | 0.005244755 |
| FDR q-value | 0.7329914 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | BIRC5 | 110408 580014 1770632 | 321 | 1.558 | 0.0208 | No | ||
| 2 | XPA | 6660133 | 423 | 1.403 | 0.0496 | No | ||
| 3 | PPP2R4 | 670280 2900097 | 1158 | 0.868 | 0.0312 | No | ||
| 4 | HPRT1 | 1690398 4920619 6350170 | 1619 | 0.663 | 0.0226 | No | ||
| 5 | MYH9 | 3120091 4850292 | 1700 | 0.628 | 0.0336 | No | ||
| 6 | ADRB2 | 3290373 | 1756 | 0.609 | 0.0455 | No | ||
| 7 | CD4 | 1090010 | 2087 | 0.501 | 0.0399 | No | ||
| 8 | ALS2 | 3130546 6400070 | 2190 | 0.474 | 0.0460 | No | ||
| 9 | ERCC5 | 770025 4540692 | 2320 | 0.436 | 0.0497 | No | ||
| 10 | TCF3 | 5080022 | 2381 | 0.418 | 0.0567 | No | ||
| 11 | NFS1 | 1170465 2680563 | 2653 | 0.353 | 0.0507 | No | ||
| 12 | BARD1 | 3170348 | 2804 | 0.323 | 0.0505 | No | ||
| 13 | APOC2 | 50100 | 2845 | 0.315 | 0.0560 | No | ||
| 14 | PVRL2 | 2760131 4050114 | 3709 | 0.188 | 0.0140 | No | ||
| 15 | MSH6 | 4480064 6520093 | 3939 | 0.166 | 0.0057 | No | ||
| 16 | TOP2A | 360717 | 3972 | 0.164 | 0.0080 | No | ||
| 17 | DSCAML1 | 1850403 5550079 | 4622 | 0.115 | -0.0243 | No | ||
| 18 | MAP3K11 | 7000039 | 4629 | 0.115 | -0.0218 | No | ||
| 19 | FOXP2 | 3520561 4150372 4760524 | 4774 | 0.107 | -0.0269 | No | ||
| 20 | AGXT | 5910338 | 4881 | 0.101 | -0.0302 | No | ||
| 21 | PVRL3 | 1400324 4200593 6650128 | 4994 | 0.095 | -0.0339 | No | ||
| 22 | VWF | 4670519 | 5064 | 0.092 | -0.0354 | No | ||
| 23 | CADM3 | 6660072 | 5099 | 0.091 | -0.0350 | No | ||
| 24 | MAFA | 70519 | 5271 | 0.084 | -0.0422 | No | ||
| 25 | APOA4 | 4120451 | 5583 | 0.072 | -0.0573 | No | ||
| 26 | IL17F | 670577 | 5850 | 0.062 | -0.0701 | No | ||
| 27 | HPS4 | 6940110 | 5971 | 0.059 | -0.0752 | No | ||
| 28 | S100B | 6520088 | 6331 | 0.049 | -0.0934 | No | ||
| 29 | GLA | 4610364 | 6348 | 0.049 | -0.0930 | No | ||
| 30 | TBX1 | 3710133 6590121 | 6391 | 0.048 | -0.0941 | No | ||
| 31 | EEA1 | 3940044 4010301 | 6875 | 0.038 | -0.1193 | No | ||
| 32 | CDA | 1980605 | 6932 | 0.037 | -0.1214 | No | ||
| 33 | SDCBP2 | 1400014 3840037 | 7230 | 0.031 | -0.1367 | No | ||
| 34 | ADRB3 | 6900072 | 7264 | 0.030 | -0.1378 | No | ||
| 35 | SLC9A7 | 5360301 | 7472 | 0.026 | -0.1483 | No | ||
| 36 | GDNF | 4050487 5720739 | 7818 | 0.021 | -0.1665 | No | ||
| 37 | MSH3 | 6770575 | 7897 | 0.020 | -0.1702 | No | ||
| 38 | GZMA | 3390315 | 7962 | 0.018 | -0.1732 | No | ||
| 39 | APOA2 | 6510364 6860411 | 7968 | 0.018 | -0.1730 | No | ||
| 40 | TYRP1 | 3170450 5690204 6290092 | 8615 | 0.009 | -0.2077 | No | ||
| 41 | NR6A1 | 4010347 | 8616 | 0.009 | -0.2075 | No | ||
| 42 | CR2 | 2850332 6450397 | 9000 | 0.003 | -0.2281 | No | ||
| 43 | NLRC4 | 5570332 | 9146 | 0.002 | -0.2359 | No | ||
| 44 | CENPF | 5050088 | 9318 | -0.001 | -0.2451 | No | ||
| 45 | MAP3K9 | 3290707 | 9692 | -0.006 | -0.2651 | No | ||
| 46 | GYS2 | 2570736 | 9740 | -0.007 | -0.2675 | No | ||
| 47 | CUBN | 630338 | 9939 | -0.010 | -0.2780 | No | ||
| 48 | ANXA9 | 1190195 | 10213 | -0.014 | -0.2924 | No | ||
| 49 | ADAM10 | 3780156 | 10357 | -0.015 | -0.2997 | No | ||
| 50 | SNX6 | 6200086 | 10390 | -0.016 | -0.3011 | No | ||
| 51 | MAP3K12 | 2470373 6420162 | 10636 | -0.020 | -0.3138 | No | ||
| 52 | CIDEA | 4560020 | 10797 | -0.022 | -0.3219 | No | ||
| 53 | TPD52 | 670463 4610541 | 10814 | -0.022 | -0.3223 | No | ||
| 54 | HEXB | 5860692 | 11092 | -0.027 | -0.3366 | No | ||
| 55 | IGSF1 | 630270 1400438 1500524 2230465 | 11461 | -0.033 | -0.3556 | No | ||
| 56 | BNIP3 | 3140270 | 11594 | -0.036 | -0.3619 | No | ||
| 57 | MAP3K5 | 6020041 6380162 | 11712 | -0.038 | -0.3673 | No | ||
| 58 | MAP3K13 | 3190017 | 11785 | -0.040 | -0.3702 | No | ||
| 59 | NPM1 | 4730427 | 12115 | -0.046 | -0.3869 | No | ||
| 60 | CAV2 | 5130286 5130563 | 12357 | -0.052 | -0.3986 | No | ||
| 61 | TYR | 5690139 | 12395 | -0.053 | -0.3993 | No | ||
| 62 | CHRNA7 | 2970446 | 12783 | -0.064 | -0.4187 | No | ||
| 63 | ABCG2 | 6370056 | 13261 | -0.079 | -0.4425 | No | ||
| 64 | ACTN3 | 3140541 6480598 | 13406 | -0.085 | -0.4482 | No | ||
| 65 | MOAP1 | 580537 | 13630 | -0.094 | -0.4580 | No | ||
| 66 | PVRL1 | 2570167 5220519 5890500 3190309 | 13799 | -0.101 | -0.4646 | No | ||
| 67 | S100A11 | 2260064 | 13805 | -0.102 | -0.4623 | No | ||
| 68 | HSP90AA1 | 4560041 5220133 2120722 | 13851 | -0.104 | -0.4622 | No | ||
| 69 | TGFB2 | 4920292 | 13877 | -0.105 | -0.4610 | No | ||
| 70 | CLIP1 | 2850162 6770397 | 14005 | -0.113 | -0.4651 | No | ||
| 71 | AOC3 | 6840129 | 14334 | -0.135 | -0.4795 | No | ||
| 72 | RRAGA | 6400603 | 14371 | -0.138 | -0.4781 | No | ||
| 73 | BCL2 | 730132 1570736 2470138 3800044 4810037 5690068 5860504 6650164 | 14485 | -0.146 | -0.4806 | No | ||
| 74 | TREX1 | 3450040 7100692 | 14646 | -0.161 | -0.4853 | No | ||
| 75 | GOLGA5 | 6770301 | 14755 | -0.170 | -0.4870 | No | ||
| 76 | SH3GLB1 | 4590079 5670187 | 14816 | -0.175 | -0.4860 | No | ||
| 77 | TPD52L1 | 4590711 5570575 | 15102 | -0.210 | -0.4962 | No | ||
| 78 | S100A1 | 6770707 | 15507 | -0.274 | -0.5114 | No | ||
| 79 | FOXP3 | 940707 1850692 | 15731 | -0.319 | -0.5156 | Yes | ||
| 80 | CADM1 | 6620128 | 15773 | -0.328 | -0.5098 | Yes | ||
| 81 | HPGD | 6770192 | 15892 | -0.355 | -0.5075 | Yes | ||
| 82 | TRPC4AP | 840520 1660402 | 15908 | -0.359 | -0.4995 | Yes | ||
| 83 | ABCG4 | 3440458 | 15928 | -0.362 | -0.4917 | Yes | ||
| 84 | MAP3K10 | 3360168 4850180 | 15998 | -0.381 | -0.4861 | Yes | ||
| 85 | TSC2 | 1450494 | 16265 | -0.452 | -0.4894 | Yes | ||
| 86 | SP1 | 6590017 | 16380 | -0.490 | -0.4836 | Yes | ||
| 87 | ABAT | 1940520 2690021 5690167 | 16421 | -0.501 | -0.4735 | Yes | ||
| 88 | BNIP3L | 1940347 | 16432 | -0.503 | -0.4617 | Yes | ||
| 89 | LDB1 | 5270601 | 16462 | -0.512 | -0.4508 | Yes | ||
| 90 | ZBTB16 | 3290022 | 16511 | -0.531 | -0.4404 | Yes | ||
| 91 | ATPIF1 | 7050577 | 16623 | -0.570 | -0.4324 | Yes | ||
| 92 | BAX | 3830008 | 16778 | -0.628 | -0.4254 | Yes | ||
| 93 | SMAD3 | 6450671 | 17125 | -0.778 | -0.4250 | Yes | ||
| 94 | MSH2 | 6180273 | 17144 | -0.787 | -0.4068 | Yes | ||
| 95 | MYO9B | 1230435 3060358 | 17207 | -0.822 | -0.3900 | Yes | ||
| 96 | VAPB | 1660451 6130333 | 17236 | -0.835 | -0.3711 | Yes | ||
| 97 | SCYE1 | 4730736 | 17288 | -0.857 | -0.3529 | Yes | ||
| 98 | S100A6 | 1690204 | 17636 | -1.028 | -0.3465 | Yes | ||
| 99 | SMAD4 | 5670519 | 17673 | -1.053 | -0.3227 | Yes | ||
| 100 | PLOD1 | 6040427 | 17854 | -1.188 | -0.3033 | Yes | ||
| 101 | EIF2AK1 | 2470301 | 17936 | -1.246 | -0.2772 | Yes | ||
| 102 | INHBB | 2680092 | 17940 | -1.248 | -0.2469 | Yes | ||
| 103 | PYCARD | 2100750 | 18005 | -1.303 | -0.2185 | Yes | ||
| 104 | STK4 | 2640152 | 18025 | -1.319 | -0.1872 | Yes | ||
| 105 | ACTN4 | 3840301 4590390 7050132 | 18044 | -1.337 | -0.1555 | Yes | ||
| 106 | ABCG1 | 60692 | 18129 | -1.423 | -0.1253 | Yes | ||
| 107 | NRBP1 | 1740750 | 18289 | -1.636 | -0.0938 | Yes | ||
| 108 | APOE | 4200671 | 18507 | -2.229 | -0.0510 | Yes | ||
| 109 | CST3 | 7050452 | 18535 | -2.325 | 0.0044 | Yes |

