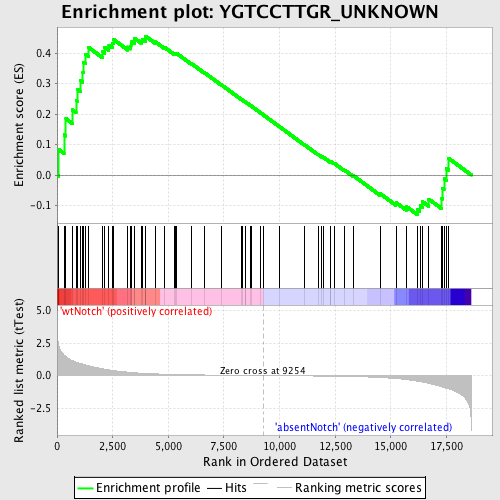
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #wtNotch_versus_absentNotch |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#wtNotch_versus_absentNotch |
| Upregulated in class | wtNotch |
| GeneSet | YGTCCTTGR_UNKNOWN |
| Enrichment Score (ES) | 0.45580712 |
| Normalized Enrichment Score (NES) | 1.2719568 |
| Nominal p-value | 0.08033827 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ALDOA | 6290672 | 72 | 2.228 | 0.0834 | Yes | ||
| 2 | KCNAB3 | 2470725 | 313 | 1.577 | 0.1323 | Yes | ||
| 3 | IGSF3 | 6450524 | 396 | 1.447 | 0.1846 | Yes | ||
| 4 | VAMP2 | 2100100 | 690 | 1.144 | 0.2137 | Yes | ||
| 5 | BAZ2A | 730184 | 852 | 1.024 | 0.2451 | Yes | ||
| 6 | OTX2 | 1190471 | 932 | 0.992 | 0.2797 | Yes | ||
| 7 | HDAC3 | 4060072 | 1049 | 0.927 | 0.3098 | Yes | ||
| 8 | PPP2R4 | 670280 2900097 | 1158 | 0.868 | 0.3380 | Yes | ||
| 9 | HSPBAP1 | 4060022 | 1188 | 0.849 | 0.3697 | Yes | ||
| 10 | YWHAG | 3780341 | 1275 | 0.804 | 0.3966 | Yes | ||
| 11 | CTRL | 2120301 | 1404 | 0.753 | 0.4192 | Yes | ||
| 12 | SYT13 | 5910110 | 2056 | 0.513 | 0.4042 | Yes | ||
| 13 | ADAM11 | 1050008 3130494 | 2140 | 0.486 | 0.4188 | Yes | ||
| 14 | ESM1 | 1410594 | 2329 | 0.434 | 0.4257 | Yes | ||
| 15 | CENTD1 | 2570592 | 2482 | 0.391 | 0.4328 | Yes | ||
| 16 | STIM1 | 6380138 | 2532 | 0.379 | 0.4450 | Yes | ||
| 17 | CRAT | 540020 2060364 3520148 | 3181 | 0.256 | 0.4201 | Yes | ||
| 18 | SNTA1 | 4570400 | 3304 | 0.238 | 0.4228 | Yes | ||
| 19 | RGL1 | 1500097 6900450 | 3322 | 0.235 | 0.4312 | Yes | ||
| 20 | DIRC2 | 2810494 | 3362 | 0.230 | 0.4381 | Yes | ||
| 21 | CYCS | 2030072 | 3475 | 0.216 | 0.4405 | Yes | ||
| 22 | ATP2A2 | 1090075 3990279 | 3485 | 0.214 | 0.4484 | Yes | ||
| 23 | PANK3 | 3060687 | 3776 | 0.182 | 0.4399 | Yes | ||
| 24 | ARMET | 5130670 | 3816 | 0.178 | 0.4448 | Yes | ||
| 25 | FHL2 | 1400008 | 3965 | 0.165 | 0.4432 | Yes | ||
| 26 | TNKS1BP1 | 7040184 | 3970 | 0.164 | 0.4495 | Yes | ||
| 27 | TMEM16D | 2650324 | 3973 | 0.164 | 0.4558 | Yes | ||
| 28 | PRKAG1 | 6450091 | 4420 | 0.128 | 0.4368 | No | ||
| 29 | SOCS5 | 3830398 7100093 | 4848 | 0.103 | 0.4178 | No | ||
| 30 | AFP | 6590358 | 5282 | 0.083 | 0.3977 | No | ||
| 31 | HRH3 | 6180040 | 5302 | 0.082 | 0.3999 | No | ||
| 32 | PHYHIPL | 2360706 3840692 | 5373 | 0.079 | 0.3993 | No | ||
| 33 | CLCNKA | 1740050 | 6040 | 0.057 | 0.3656 | No | ||
| 34 | CBX4 | 70332 | 6634 | 0.043 | 0.3353 | No | ||
| 35 | ARPC5 | 1740411 3450300 | 7402 | 0.027 | 0.2950 | No | ||
| 36 | NPTX2 | 70021 | 8277 | 0.014 | 0.2484 | No | ||
| 37 | SYT7 | 2450528 3390500 3610687 | 8328 | 0.013 | 0.2463 | No | ||
| 38 | AMY2A | 580138 | 8467 | 0.011 | 0.2393 | No | ||
| 39 | ELAVL2 | 360181 | 8471 | 0.011 | 0.2395 | No | ||
| 40 | EMX2 | 1660092 | 8690 | 0.008 | 0.2281 | No | ||
| 41 | TRIM2 | 430546 1170286 | 8732 | 0.007 | 0.2261 | No | ||
| 42 | MLLT6 | 3870168 | 9126 | 0.002 | 0.2050 | No | ||
| 43 | PAM | 5290528 | 9287 | -0.000 | 0.1964 | No | ||
| 44 | SNCB | 3360133 | 9986 | -0.010 | 0.1592 | No | ||
| 45 | KCNJ16 | 4780273 | 11097 | -0.027 | 0.1004 | No | ||
| 46 | HOXD11 | 2260242 | 11735 | -0.038 | 0.0676 | No | ||
| 47 | DRD3 | 4780402 | 11897 | -0.041 | 0.0605 | No | ||
| 48 | MT3 | 1450537 | 11985 | -0.043 | 0.0575 | No | ||
| 49 | SEMA6D | 4050324 5860138 6350307 | 12295 | -0.051 | 0.0429 | No | ||
| 50 | CHST8 | 1940050 | 12299 | -0.051 | 0.0447 | No | ||
| 51 | PRKCE | 5700053 | 12458 | -0.054 | 0.0383 | No | ||
| 52 | ZIC3 | 380020 | 12924 | -0.068 | 0.0159 | No | ||
| 53 | GABRA3 | 4810142 | 13301 | -0.080 | -0.0012 | No | ||
| 54 | PRKACA | 2640731 4050048 | 14523 | -0.149 | -0.0612 | No | ||
| 55 | SYNPO2L | 5390719 | 15232 | -0.228 | -0.0904 | No | ||
| 56 | ATP5B | 3870138 | 15689 | -0.308 | -0.1029 | No | ||
| 57 | THRA | 6770341 110164 1990600 | 16183 | -0.427 | -0.1128 | No | ||
| 58 | CS | 5080600 | 16311 | -0.469 | -0.1012 | No | ||
| 59 | CITED2 | 5670114 5130088 | 16426 | -0.502 | -0.0877 | No | ||
| 60 | HIST1H1A | 1450131 | 16703 | -0.600 | -0.0791 | No | ||
| 61 | HMGCS1 | 6620452 | 17295 | -0.860 | -0.0772 | No | ||
| 62 | H2AFZ | 1470168 | 17329 | -0.879 | -0.0445 | No | ||
| 63 | MCAM | 5130270 6620577 | 17393 | -0.917 | -0.0120 | No | ||
| 64 | GAS6 | 4480021 | 17500 | -0.979 | 0.0207 | No | ||
| 65 | SLC38A2 | 1470242 3800026 | 17587 | -1.006 | 0.0555 | No |

