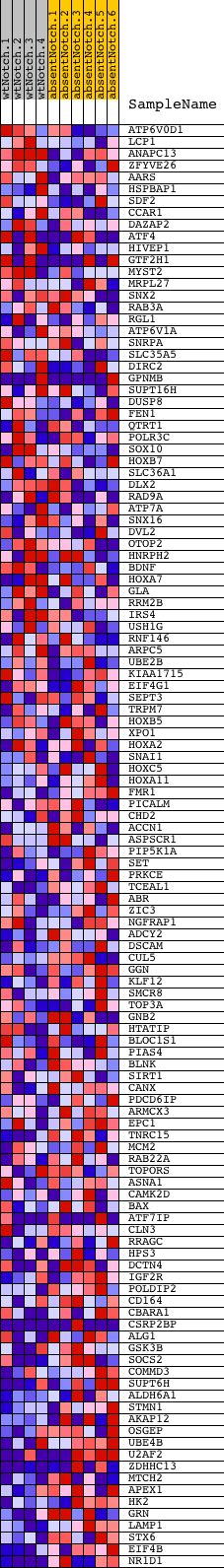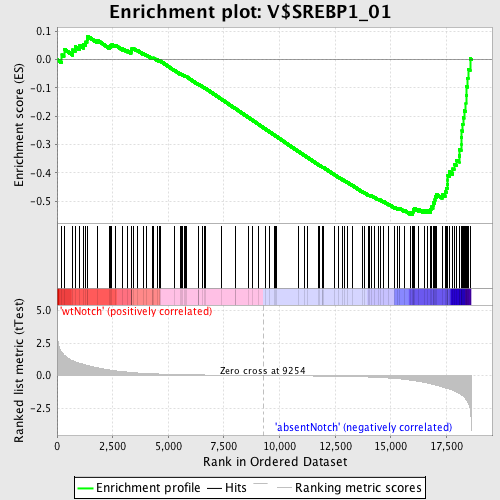
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #wtNotch_versus_absentNotch |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#wtNotch_versus_absentNotch |
| Upregulated in class | absentNotch |
| GeneSet | V$SREBP1_01 |
| Enrichment Score (ES) | -0.54709315 |
| Normalized Enrichment Score (NES) | -1.5849336 |
| Nominal p-value | 0.0017921147 |
| FDR q-value | 0.16027342 |
| FWER p-Value | 0.532 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ATP6V0D1 | 360348 5900021 6290605 | 218 | 1.752 | 0.0166 | No | ||
| 2 | LCP1 | 4280397 | 310 | 1.578 | 0.0373 | No | ||
| 3 | ANAPC13 | 3800446 | 706 | 1.133 | 0.0343 | No | ||
| 4 | ZFYVE26 | 6860152 | 810 | 1.060 | 0.0459 | No | ||
| 5 | AARS | 1570279 2370136 | 1014 | 0.940 | 0.0501 | No | ||
| 6 | HSPBAP1 | 4060022 | 1188 | 0.849 | 0.0545 | No | ||
| 7 | SDF2 | 2690601 | 1284 | 0.800 | 0.0624 | No | ||
| 8 | CCAR1 | 3190092 | 1344 | 0.777 | 0.0718 | No | ||
| 9 | DAZAP2 | 6200102 | 1366 | 0.770 | 0.0831 | No | ||
| 10 | ATF4 | 730441 1740195 | 1813 | 0.593 | 0.0686 | No | ||
| 11 | HIVEP1 | 3850632 | 2376 | 0.420 | 0.0450 | No | ||
| 12 | GTF2H1 | 2570746 2650148 | 2403 | 0.412 | 0.0503 | No | ||
| 13 | MYST2 | 4540494 | 2462 | 0.398 | 0.0536 | No | ||
| 14 | MRPL27 | 2060131 | 2609 | 0.363 | 0.0516 | No | ||
| 15 | SNX2 | 2470068 3360364 6510064 | 2944 | 0.298 | 0.0384 | No | ||
| 16 | RAB3A | 3940288 | 3158 | 0.259 | 0.0311 | No | ||
| 17 | RGL1 | 1500097 6900450 | 3322 | 0.235 | 0.0261 | No | ||
| 18 | ATP6V1A | 6590242 | 3325 | 0.234 | 0.0297 | No | ||
| 19 | SNRPA | 2570100 4150121 6940484 | 3329 | 0.234 | 0.0334 | No | ||
| 20 | SLC35A5 | 2970670 4480546 | 3358 | 0.231 | 0.0356 | No | ||
| 21 | DIRC2 | 2810494 | 3362 | 0.230 | 0.0392 | No | ||
| 22 | GPNMB | 1780138 2340176 3390403 4560156 | 3431 | 0.221 | 0.0391 | No | ||
| 23 | SUPT16H | 1240333 | 3594 | 0.200 | 0.0336 | No | ||
| 24 | DUSP8 | 6650039 | 3886 | 0.171 | 0.0206 | No | ||
| 25 | FEN1 | 1770541 | 4018 | 0.160 | 0.0161 | No | ||
| 26 | QTRT1 | 1340162 | 4267 | 0.139 | 0.0050 | No | ||
| 27 | POLR3C | 6940546 | 4305 | 0.136 | 0.0052 | No | ||
| 28 | SOX10 | 6200538 | 4333 | 0.134 | 0.0059 | No | ||
| 29 | HOXB7 | 4540706 4610358 | 4493 | 0.123 | -0.0007 | No | ||
| 30 | SLC36A1 | 5080242 | 4620 | 0.116 | -0.0057 | No | ||
| 31 | DLX2 | 2320438 4200673 | 4635 | 0.115 | -0.0046 | No | ||
| 32 | RAD9A | 110300 1940632 3990390 6040014 | 5268 | 0.084 | -0.0374 | No | ||
| 33 | ATP7A | 6550168 | 5531 | 0.073 | -0.0504 | No | ||
| 34 | SNX16 | 870446 | 5571 | 0.072 | -0.0513 | No | ||
| 35 | DVL2 | 6110162 | 5617 | 0.070 | -0.0526 | No | ||
| 36 | OTOP2 | 610068 | 5730 | 0.066 | -0.0576 | No | ||
| 37 | HNRPH2 | 2630040 5720671 | 5769 | 0.065 | -0.0586 | No | ||
| 38 | BDNF | 2940128 3520368 | 5794 | 0.064 | -0.0588 | No | ||
| 39 | HOXA7 | 5910152 | 6346 | 0.049 | -0.0878 | No | ||
| 40 | GLA | 4610364 | 6348 | 0.049 | -0.0871 | No | ||
| 41 | RRM2B | 1690025 1690465 4010400 | 6369 | 0.048 | -0.0874 | No | ||
| 42 | IRS4 | 430341 1190450 | 6513 | 0.045 | -0.0944 | No | ||
| 43 | USH1G | 2940446 | 6630 | 0.043 | -0.1000 | No | ||
| 44 | RNF146 | 1660735 | 6664 | 0.042 | -0.1011 | No | ||
| 45 | ARPC5 | 1740411 3450300 | 7402 | 0.027 | -0.1405 | No | ||
| 46 | UBE2B | 4780497 | 8000 | 0.018 | -0.1725 | No | ||
| 47 | KIAA1715 | 1190551 | 8014 | 0.018 | -0.1729 | No | ||
| 48 | EIF4G1 | 4070446 | 8585 | 0.009 | -0.2036 | No | ||
| 49 | SEPT3 | 3830022 5290079 | 8771 | 0.006 | -0.2135 | No | ||
| 50 | TRPM7 | 2510408 | 9040 | 0.003 | -0.2279 | No | ||
| 51 | HOXB5 | 2900538 | 9344 | -0.001 | -0.2443 | No | ||
| 52 | XPO1 | 540707 | 9544 | -0.004 | -0.2550 | No | ||
| 53 | HOXA2 | 2120121 | 9777 | -0.007 | -0.2674 | No | ||
| 54 | SNAI1 | 6590253 | 9796 | -0.007 | -0.2683 | No | ||
| 55 | HOXC5 | 1580017 | 9850 | -0.008 | -0.2710 | No | ||
| 56 | HOXA11 | 6980133 | 10835 | -0.023 | -0.3238 | No | ||
| 57 | FMR1 | 5050075 | 11131 | -0.028 | -0.3393 | No | ||
| 58 | PICALM | 1940280 | 11250 | -0.030 | -0.3452 | No | ||
| 59 | CHD2 | 4070039 | 11727 | -0.038 | -0.3703 | No | ||
| 60 | ACCN1 | 2060139 2190541 | 11793 | -0.040 | -0.3732 | No | ||
| 61 | ASPSCR1 | 610167 3990739 | 11927 | -0.042 | -0.3797 | No | ||
| 62 | PIP5K1A | 4560672 | 11935 | -0.042 | -0.3794 | No | ||
| 63 | SET | 6650286 | 11986 | -0.043 | -0.3814 | No | ||
| 64 | PRKCE | 5700053 | 12458 | -0.054 | -0.4060 | No | ||
| 65 | TCEAL1 | 870577 | 12651 | -0.059 | -0.4154 | No | ||
| 66 | ABR | 610079 1170609 3610195 5670050 | 12839 | -0.065 | -0.4245 | No | ||
| 67 | ZIC3 | 380020 | 12924 | -0.068 | -0.4279 | No | ||
| 68 | NGFRAP1 | 6770195 | 13031 | -0.071 | -0.4325 | No | ||
| 69 | ADCY2 | 4610100 | 13282 | -0.080 | -0.4447 | No | ||
| 70 | DSCAM | 1780050 2450731 2810438 | 13725 | -0.098 | -0.4670 | No | ||
| 71 | CUL5 | 450142 | 13798 | -0.101 | -0.4693 | No | ||
| 72 | GGN | 1410528 4210082 5290170 | 14011 | -0.113 | -0.4789 | No | ||
| 73 | KLF12 | 1660095 4810288 5340546 6520286 | 14054 | -0.117 | -0.4793 | No | ||
| 74 | SMCR8 | 730653 | 14117 | -0.120 | -0.4807 | No | ||
| 75 | TOP3A | 7000435 | 14254 | -0.129 | -0.4860 | No | ||
| 76 | GNB2 | 2350053 | 14436 | -0.142 | -0.4934 | No | ||
| 77 | HTATIP | 5360739 | 14522 | -0.149 | -0.4956 | No | ||
| 78 | BLOC1S1 | 130341 3940717 | 14668 | -0.163 | -0.5008 | No | ||
| 79 | PIAS4 | 6040113 | 14906 | -0.187 | -0.5106 | No | ||
| 80 | BLNK | 70288 6590092 | 15184 | -0.222 | -0.5220 | No | ||
| 81 | SIRT1 | 1190731 | 15314 | -0.239 | -0.5251 | No | ||
| 82 | CANX | 4760402 | 15396 | -0.252 | -0.5254 | No | ||
| 83 | PDCD6IP | 1990403 | 15613 | -0.293 | -0.5323 | No | ||
| 84 | ARMCX3 | 3290309 | 15873 | -0.349 | -0.5407 | No | ||
| 85 | EPC1 | 1570050 5290095 6900193 | 15993 | -0.380 | -0.5409 | Yes | ||
| 86 | TNRC15 | 6760746 | 16000 | -0.382 | -0.5351 | Yes | ||
| 87 | MCM2 | 5050139 | 16009 | -0.385 | -0.5293 | Yes | ||
| 88 | RAB22A | 110500 3830707 | 16058 | -0.397 | -0.5254 | Yes | ||
| 89 | TOPORS | 4060592 | 16243 | -0.443 | -0.5282 | Yes | ||
| 90 | ASNA1 | 3830041 | 16495 | -0.524 | -0.5333 | Yes | ||
| 91 | CAMK2D | 2370685 | 16654 | -0.579 | -0.5324 | Yes | ||
| 92 | BAX | 3830008 | 16778 | -0.628 | -0.5289 | Yes | ||
| 93 | ATF7IP | 2690176 6770021 | 16813 | -0.642 | -0.5203 | Yes | ||
| 94 | CLN3 | 780368 | 16896 | -0.678 | -0.5138 | Yes | ||
| 95 | RRAGC | 5910348 | 16905 | -0.681 | -0.5032 | Yes | ||
| 96 | HPS3 | 6650348 | 16981 | -0.711 | -0.4957 | Yes | ||
| 97 | DCTN4 | 3390010 5550170 | 16989 | -0.716 | -0.4845 | Yes | ||
| 98 | IGF2R | 1570402 | 17052 | -0.749 | -0.4757 | Yes | ||
| 99 | POLDIP2 | 2650139 | 17303 | -0.864 | -0.4752 | Yes | ||
| 100 | CD164 | 3830594 | 17441 | -0.949 | -0.4672 | Yes | ||
| 101 | CBARA1 | 360471 | 17489 | -0.973 | -0.4540 | Yes | ||
| 102 | CSRP2BP | 4570717 5550333 | 17534 | -0.996 | -0.4402 | Yes | ||
| 103 | ALG1 | 2570112 | 17557 | -0.999 | -0.4252 | Yes | ||
| 104 | GSK3B | 5360348 | 17561 | -1.000 | -0.4092 | Yes | ||
| 105 | SOCS2 | 4760692 | 17628 | -1.024 | -0.3962 | Yes | ||
| 106 | COMMD3 | 2510259 | 17788 | -1.142 | -0.3863 | Yes | ||
| 107 | SUPT6H | 3170082 | 17843 | -1.182 | -0.3700 | Yes | ||
| 108 | ALDH6A1 | 1770746 | 17943 | -1.249 | -0.3551 | Yes | ||
| 109 | STMN1 | 1990717 | 18068 | -1.364 | -0.3397 | Yes | ||
| 110 | AKAP12 | 1450739 | 18094 | -1.383 | -0.3187 | Yes | ||
| 111 | OSGEP | 5700239 | 18158 | -1.465 | -0.2983 | Yes | ||
| 112 | UBE4B | 780008 3610154 | 18163 | -1.473 | -0.2747 | Yes | ||
| 113 | U2AF2 | 450600 4670128 5420292 | 18196 | -1.513 | -0.2519 | Yes | ||
| 114 | ZDHHC13 | 430170 4230021 | 18216 | -1.538 | -0.2280 | Yes | ||
| 115 | MTCH2 | 3780603 6620215 | 18248 | -1.571 | -0.2042 | Yes | ||
| 116 | APEX1 | 3190519 | 18290 | -1.637 | -0.1799 | Yes | ||
| 117 | HK2 | 2640722 | 18377 | -1.825 | -0.1549 | Yes | ||
| 118 | GRN | 4050239 | 18380 | -1.839 | -0.1252 | Yes | ||
| 119 | LAMP1 | 2470524 | 18413 | -1.920 | -0.0959 | Yes | ||
| 120 | STX6 | 4850750 6770278 | 18434 | -1.963 | -0.0651 | Yes | ||
| 121 | EIF4B | 5390563 5270577 | 18473 | -2.090 | -0.0333 | Yes | ||
| 122 | NR1D1 | 2360471 770746 6590204 | 18562 | -2.528 | 0.0029 | Yes |