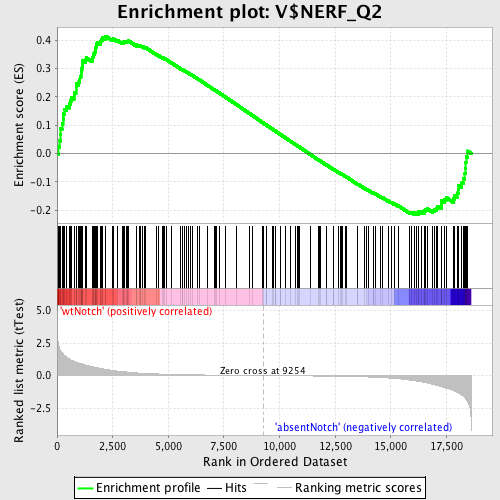
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #wtNotch_versus_absentNotch |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#wtNotch_versus_absentNotch |
| Upregulated in class | wtNotch |
| GeneSet | V$NERF_Q2 |
| Enrichment Score (ES) | 0.41497055 |
| Normalized Enrichment Score (NES) | 1.2872821 |
| Nominal p-value | 0.03217822 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RASAL1 | 3710112 | 57 | 2.379 | 0.0246 | Yes | ||
| 2 | SH2D2A | 2970594 | 126 | 2.034 | 0.0446 | Yes | ||
| 3 | ZFPM1 | 6760093 | 137 | 2.005 | 0.0674 | Yes | ||
| 4 | FBXO36 | 1410086 | 156 | 1.921 | 0.0888 | Yes | ||
| 5 | UBE2L3 | 1690707 5900242 | 222 | 1.745 | 0.1056 | Yes | ||
| 6 | FGR | 3780047 | 265 | 1.662 | 0.1227 | Yes | ||
| 7 | HMGA1 | 6580408 | 293 | 1.609 | 0.1400 | Yes | ||
| 8 | DEF6 | 840593 | 324 | 1.555 | 0.1565 | Yes | ||
| 9 | CREB3 | 5220408 | 428 | 1.396 | 0.1672 | Yes | ||
| 10 | FBS1 | 2570520 | 547 | 1.269 | 0.1756 | Yes | ||
| 11 | PPP1R9B | 3130619 | 585 | 1.227 | 0.1878 | Yes | ||
| 12 | TPP2 | 3850093 | 631 | 1.190 | 0.1993 | Yes | ||
| 13 | HMGN2 | 3140091 | 762 | 1.096 | 0.2050 | Yes | ||
| 14 | JUNB | 4230048 | 795 | 1.073 | 0.2158 | Yes | ||
| 15 | POLD4 | 2320082 | 850 | 1.024 | 0.2248 | Yes | ||
| 16 | BAZ2A | 730184 | 852 | 1.024 | 0.2366 | Yes | ||
| 17 | RASA1 | 1240315 | 869 | 1.018 | 0.2476 | Yes | ||
| 18 | HIC1 | 2650142 | 981 | 0.963 | 0.2528 | Yes | ||
| 19 | HCLS1 | 7100309 | 1017 | 0.939 | 0.2619 | Yes | ||
| 20 | RPS6KA4 | 2190524 | 1028 | 0.934 | 0.2722 | Yes | ||
| 21 | SRPR | 6590647 6370114 | 1074 | 0.917 | 0.2804 | Yes | ||
| 22 | STAT5A | 2680458 | 1078 | 0.913 | 0.2909 | Yes | ||
| 23 | LANCL1 | 1770079 6420670 | 1108 | 0.896 | 0.2998 | Yes | ||
| 24 | PTPN6 | 4230128 | 1132 | 0.885 | 0.3088 | Yes | ||
| 25 | CORO1A | 3140609 3190020 3190037 | 1148 | 0.875 | 0.3182 | Yes | ||
| 26 | ITPR3 | 4010632 | 1153 | 0.873 | 0.3282 | Yes | ||
| 27 | MAPK3 | 580161 4780035 | 1279 | 0.803 | 0.3307 | Yes | ||
| 28 | TMEM4 | 6220091 | 1301 | 0.793 | 0.3388 | Yes | ||
| 29 | ZHX2 | 2900452 | 1580 | 0.680 | 0.3317 | Yes | ||
| 30 | CAMK1D | 1660100 2350309 | 1609 | 0.667 | 0.3380 | Yes | ||
| 31 | MCTS1 | 6130133 | 1614 | 0.666 | 0.3455 | Yes | ||
| 32 | TGIF2 | 3840364 4050520 | 1638 | 0.655 | 0.3519 | Yes | ||
| 33 | TRAPPC3 | 2030176 6020609 6590373 6760019 | 1687 | 0.633 | 0.3566 | Yes | ||
| 34 | ATP1A1 | 5670451 | 1711 | 0.623 | 0.3626 | Yes | ||
| 35 | TFRC | 4050551 | 1727 | 0.618 | 0.3690 | Yes | ||
| 36 | USP5 | 4850017 | 1734 | 0.616 | 0.3759 | Yes | ||
| 37 | SLC4A2 | 2690114 | 1773 | 0.604 | 0.3809 | Yes | ||
| 38 | RGS3 | 60670 540736 1340180 1500369 3390735 4010131 4610402 6380114 | 1782 | 0.602 | 0.3874 | Yes | ||
| 39 | PIK3CG | 5890110 | 1823 | 0.589 | 0.3921 | Yes | ||
| 40 | AGPAT1 | 610056 | 1942 | 0.551 | 0.3922 | Yes | ||
| 41 | LAT | 3170025 | 1959 | 0.545 | 0.3976 | Yes | ||
| 42 | IKBKB | 6840072 | 1993 | 0.532 | 0.4020 | Yes | ||
| 43 | RANBP1 | 430215 1090180 | 2036 | 0.519 | 0.4058 | Yes | ||
| 44 | TLN1 | 6590411 | 2043 | 0.517 | 0.4115 | Yes | ||
| 45 | EBAG9 | 5090739 7330181 | 2181 | 0.477 | 0.4096 | Yes | ||
| 46 | LYL1 | 2900242 | 2186 | 0.476 | 0.4150 | Yes | ||
| 47 | GGTL3 | 1770288 | 2478 | 0.392 | 0.4038 | No | ||
| 48 | ROM1 | 3130685 | 2513 | 0.383 | 0.4064 | No | ||
| 49 | SNCG | 3120725 | 2702 | 0.343 | 0.4002 | No | ||
| 50 | ARHGDIB | 4730433 | 2922 | 0.301 | 0.3918 | No | ||
| 51 | DDIT3 | 780373 | 2986 | 0.290 | 0.3918 | No | ||
| 52 | LYPLA2 | 1500112 | 3003 | 0.288 | 0.3943 | No | ||
| 53 | MAP4K1 | 4060692 | 3032 | 0.284 | 0.3961 | No | ||
| 54 | C1QTNF6 | 2030592 | 3126 | 0.267 | 0.3941 | No | ||
| 55 | SPRED2 | 510138 540195 | 3150 | 0.261 | 0.3959 | No | ||
| 56 | DPP3 | 2680725 3780670 | 3184 | 0.255 | 0.3971 | No | ||
| 57 | NTRK3 | 4050687 4070167 4610026 6100484 | 3216 | 0.251 | 0.3984 | No | ||
| 58 | SALL1 | 5420020 7050195 | 3563 | 0.205 | 0.3820 | No | ||
| 59 | FLT1 | 3830167 4920438 | 3565 | 0.205 | 0.3843 | No | ||
| 60 | PEX11B | 6510152 | 3701 | 0.189 | 0.3792 | No | ||
| 61 | PEX16 | 4560044 | 3728 | 0.186 | 0.3800 | No | ||
| 62 | MID1 | 4070114 4760458 | 3739 | 0.185 | 0.3816 | No | ||
| 63 | LRRFIP2 | 670575 730152 6590438 | 3850 | 0.175 | 0.3777 | No | ||
| 64 | SEMA4C | 1980315 | 3948 | 0.166 | 0.3743 | No | ||
| 65 | ERG | 50154 1770739 | 3951 | 0.166 | 0.3762 | No | ||
| 66 | CLIC1 | 110600 | 4469 | 0.125 | 0.3496 | No | ||
| 67 | NPY | 3170138 | 4568 | 0.119 | 0.3457 | No | ||
| 68 | RBBP7 | 430113 450450 2370309 | 4730 | 0.110 | 0.3382 | No | ||
| 69 | MDFI | 6200041 | 4760 | 0.108 | 0.3379 | No | ||
| 70 | MPL | 2360253 | 4837 | 0.103 | 0.3350 | No | ||
| 71 | PIGW | 4810072 | 4917 | 0.099 | 0.3318 | No | ||
| 72 | CNNM1 | 2190433 | 5143 | 0.089 | 0.3207 | No | ||
| 73 | HOXC4 | 1940193 | 5550 | 0.073 | 0.2995 | No | ||
| 74 | BMP4 | 380113 | 5651 | 0.069 | 0.2949 | No | ||
| 75 | EGR3 | 6940128 | 5654 | 0.069 | 0.2956 | No | ||
| 76 | GMFB | 6590035 | 5746 | 0.065 | 0.2914 | No | ||
| 77 | ADAMTS4 | 430435 | 5816 | 0.063 | 0.2884 | No | ||
| 78 | ROBO4 | 6040471 6650717 | 5902 | 0.061 | 0.2845 | No | ||
| 79 | PITX2 | 870537 2690139 | 6007 | 0.058 | 0.2796 | No | ||
| 80 | LYN | 6040600 | 6066 | 0.056 | 0.2771 | No | ||
| 81 | CAPZA1 | 3140390 | 6295 | 0.050 | 0.2653 | No | ||
| 82 | PLCB2 | 360132 | 6384 | 0.048 | 0.2611 | No | ||
| 83 | POU6F2 | 5270451 | 6769 | 0.040 | 0.2407 | No | ||
| 84 | DOK1 | 1050279 2680112 5220273 | 7069 | 0.034 | 0.2249 | No | ||
| 85 | OLFML2A | 5890039 | 7123 | 0.033 | 0.2224 | No | ||
| 86 | LSAMP | 2810056 6550082 | 7158 | 0.032 | 0.2210 | No | ||
| 87 | STAT5B | 6200026 | 7297 | 0.029 | 0.2138 | No | ||
| 88 | UBTF | 4210450 6840403 | 7583 | 0.025 | 0.1987 | No | ||
| 89 | ST7L | 840735 2360576 | 8066 | 0.017 | 0.1727 | No | ||
| 90 | FOSL1 | 430021 | 8666 | 0.008 | 0.1404 | No | ||
| 91 | TENC1 | 1980403 7050575 | 8799 | 0.006 | 0.1333 | No | ||
| 92 | RHOV | 5860100 | 9211 | 0.001 | 0.1110 | No | ||
| 93 | SDCCAG8 | 1780687 | 9273 | -0.000 | 0.1077 | No | ||
| 94 | NFIA | 2760129 5860278 | 9392 | -0.002 | 0.1013 | No | ||
| 95 | GRM3 | 4540242 | 9402 | -0.002 | 0.1009 | No | ||
| 96 | POU4F3 | 2690035 | 9660 | -0.005 | 0.0870 | No | ||
| 97 | CPNE8 | 7040039 | 9735 | -0.007 | 0.0831 | No | ||
| 98 | ACTR3 | 1400497 | 9805 | -0.007 | 0.0794 | No | ||
| 99 | EFNA1 | 3840672 | 9824 | -0.008 | 0.0785 | No | ||
| 100 | FCER1G | 5550020 | 10022 | -0.011 | 0.0680 | No | ||
| 101 | TWIST1 | 5130619 | 10033 | -0.011 | 0.0676 | No | ||
| 102 | NRXN3 | 1400546 3360471 | 10279 | -0.014 | 0.0545 | No | ||
| 103 | GOLGA7 | 6980121 | 10492 | -0.018 | 0.0432 | No | ||
| 104 | PAX6 | 1190025 | 10695 | -0.021 | 0.0325 | No | ||
| 105 | NLK | 2030010 2450041 | 10808 | -0.022 | 0.0267 | No | ||
| 106 | TRO | 1980113 | 10841 | -0.023 | 0.0252 | No | ||
| 107 | JAG1 | 3440390 | 10896 | -0.024 | 0.0225 | No | ||
| 108 | PTPRN | 5900577 | 11371 | -0.031 | -0.0028 | No | ||
| 109 | CHD2 | 4070039 | 11727 | -0.038 | -0.0216 | No | ||
| 110 | CNTNAP1 | 6650132 | 11783 | -0.040 | -0.0241 | No | ||
| 111 | ATOH8 | 4540204 | 11847 | -0.041 | -0.0270 | No | ||
| 112 | LOXL3 | 4010168 | 12097 | -0.046 | -0.0400 | No | ||
| 113 | ELK3 | 460377 1400762 3710497 | 12124 | -0.046 | -0.0409 | No | ||
| 114 | SHC3 | 2690398 | 12407 | -0.053 | -0.0555 | No | ||
| 115 | HOXB9 | 2640687 | 12435 | -0.054 | -0.0564 | No | ||
| 116 | BTBD12 | 2940072 | 12661 | -0.059 | -0.0679 | No | ||
| 117 | KLF13 | 4670563 | 12746 | -0.062 | -0.0717 | No | ||
| 118 | STARD13 | 540722 | 12760 | -0.063 | -0.0717 | No | ||
| 119 | MMRN2 | 1740047 | 12826 | -0.065 | -0.0745 | No | ||
| 120 | BCOR | 1660452 3940053 | 12970 | -0.069 | -0.0814 | No | ||
| 121 | PAK3 | 4210136 | 13014 | -0.071 | -0.0829 | No | ||
| 122 | TUSC3 | 1690411 | 13502 | -0.089 | -0.1083 | No | ||
| 123 | KCND1 | 6860278 | 13521 | -0.089 | -0.1082 | No | ||
| 124 | UCHL5 | 540139 1690053 | 13797 | -0.101 | -0.1219 | No | ||
| 125 | ZNF23 | 1090537 | 13920 | -0.108 | -0.1273 | No | ||
| 126 | CENTD2 | 60408 2510156 6100494 | 13988 | -0.112 | -0.1296 | No | ||
| 127 | FXYD5 | 4060301 6590400 | 14218 | -0.127 | -0.1405 | No | ||
| 128 | SH3TC1 | 3120162 | 14220 | -0.127 | -0.1391 | No | ||
| 129 | CDK5 | 940348 | 14297 | -0.133 | -0.1417 | No | ||
| 130 | FGFR2 | 4670601 5570402 6940377 | 14526 | -0.150 | -0.1523 | No | ||
| 131 | UQCRH | 4210463 | 14642 | -0.161 | -0.1567 | No | ||
| 132 | PCDH7 | 450398 4150040 | 14882 | -0.185 | -0.1675 | No | ||
| 133 | FURIN | 4120168 | 15017 | -0.199 | -0.1724 | No | ||
| 134 | SLITRK6 | 7320739 | 15142 | -0.215 | -0.1766 | No | ||
| 135 | CD79A | 3450563 | 15327 | -0.242 | -0.1838 | No | ||
| 136 | RPS3 | 1570017 2370170 | 15834 | -0.342 | -0.2072 | No | ||
| 137 | HYAL2 | 1500152 | 15921 | -0.361 | -0.2077 | No | ||
| 138 | ATP5H | 670438 | 16050 | -0.394 | -0.2100 | No | ||
| 139 | CAP1 | 2650278 | 16144 | -0.417 | -0.2102 | No | ||
| 140 | TOPORS | 4060592 | 16243 | -0.443 | -0.2104 | No | ||
| 141 | ARRB2 | 2060441 | 16245 | -0.443 | -0.2053 | No | ||
| 142 | TCF12 | 3610324 7000156 | 16359 | -0.484 | -0.2058 | No | ||
| 143 | CD79B | 1450066 3390358 | 16503 | -0.528 | -0.2074 | No | ||
| 144 | ITPKB | 2900195 7100039 | 16516 | -0.533 | -0.2018 | No | ||
| 145 | DPP8 | 3120136 | 16548 | -0.545 | -0.1971 | No | ||
| 146 | FKBP10 | 1340706 | 16635 | -0.572 | -0.1951 | No | ||
| 147 | MYOHD1 | 1410520 | 16882 | -0.673 | -0.2006 | No | ||
| 148 | CDCA3 | 3940397 | 16976 | -0.709 | -0.1974 | No | ||
| 149 | GPR132 | 4150114 | 17061 | -0.752 | -0.1932 | No | ||
| 150 | MAP2K6 | 1230056 2940204 | 17105 | -0.770 | -0.1866 | No | ||
| 151 | SLC30A7 | 2230463 | 17267 | -0.848 | -0.1854 | No | ||
| 152 | CALM2 | 6620463 | 17272 | -0.852 | -0.1757 | No | ||
| 153 | SECISBP2 | 6520309 | 17285 | -0.856 | -0.1664 | No | ||
| 154 | STARD8 | 1780047 | 17407 | -0.929 | -0.1621 | No | ||
| 155 | NDUFS2 | 4850020 6200402 | 17485 | -0.970 | -0.1550 | No | ||
| 156 | DIABLO | 2360170 | 17820 | -1.164 | -0.1595 | No | ||
| 157 | HTF9C | 3190048 4120132 | 17852 | -1.188 | -0.1474 | No | ||
| 158 | FREQ | 3140039 4060600 4560670 | 17980 | -1.275 | -0.1394 | No | ||
| 159 | NFATC3 | 5050520 | 18034 | -1.327 | -0.1268 | No | ||
| 160 | SLCO2B1 | 3450019 | 18045 | -1.338 | -0.1118 | No | ||
| 161 | ELOVL5 | 3800170 | 18194 | -1.511 | -0.1022 | No | ||
| 162 | EXTL2 | 5050433 | 18262 | -1.585 | -0.0874 | No | ||
| 163 | LST1 | 5420372 | 18328 | -1.714 | -0.0709 | No | ||
| 164 | VCL | 4120487 | 18343 | -1.734 | -0.0515 | No | ||
| 165 | ARHGAP8 | 380333 5390487 | 18372 | -1.806 | -0.0320 | No | ||
| 166 | HN1 | 3360156 | 18382 | -1.852 | -0.0109 | No | ||
| 167 | FCGR2B | 780750 | 18452 | -2.020 | 0.0089 | No |

