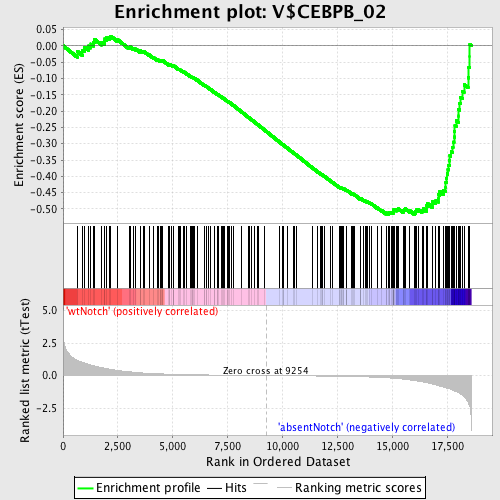
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #wtNotch_versus_absentNotch |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#wtNotch_versus_absentNotch |
| Upregulated in class | absentNotch |
| GeneSet | V$CEBPB_02 |
| Enrichment Score (ES) | -0.5168637 |
| Normalized Enrichment Score (NES) | -1.5747246 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.15887272 |
| FWER p-Value | 0.579 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SLC25A12 | 7400731 | 640 | 1.185 | -0.0166 | No | ||
| 2 | RAB3IP | 5550100 | 878 | 1.009 | -0.0140 | No | ||
| 3 | NDRG1 | 1340040 | 965 | 0.975 | -0.0038 | No | ||
| 4 | ITPR3 | 4010632 | 1153 | 0.873 | -0.0006 | No | ||
| 5 | GOT1 | 2940465 | 1270 | 0.808 | 0.0055 | No | ||
| 6 | RIN3 | 1940044 | 1390 | 0.758 | 0.0106 | No | ||
| 7 | SMARCAD1 | 5390619 | 1425 | 0.743 | 0.0201 | No | ||
| 8 | ADRB2 | 3290373 | 1756 | 0.609 | 0.0116 | No | ||
| 9 | CUEDC1 | 1660463 | 1893 | 0.566 | 0.0128 | No | ||
| 10 | PDE3B | 2030563 | 1905 | 0.562 | 0.0208 | No | ||
| 11 | CDKN1B | 3800025 6450044 | 1990 | 0.533 | 0.0244 | No | ||
| 12 | DLX1 | 360168 | 2109 | 0.493 | 0.0256 | No | ||
| 13 | STAT3 | 460040 3710341 | 2171 | 0.480 | 0.0296 | No | ||
| 14 | TGFB3 | 1070041 | 2466 | 0.396 | 0.0197 | No | ||
| 15 | MYO1C | 3190398 | 3011 | 0.287 | -0.0054 | No | ||
| 16 | PRG4 | 5860156 6650086 | 3050 | 0.280 | -0.0032 | No | ||
| 17 | SEPHS2 | 6770446 | 3203 | 0.253 | -0.0076 | No | ||
| 18 | RG9MTD2 | 6520100 | 3309 | 0.237 | -0.0097 | No | ||
| 19 | WNT5A | 840685 3120152 | 3512 | 0.211 | -0.0174 | No | ||
| 20 | BCL11A | 6860369 | 3523 | 0.209 | -0.0147 | No | ||
| 21 | BMF | 610102 | 3658 | 0.193 | -0.0190 | No | ||
| 22 | FBXW7 | 4210338 7050280 | 3691 | 0.190 | -0.0179 | No | ||
| 23 | ERF | 110551 1770278 | 3922 | 0.168 | -0.0278 | No | ||
| 24 | ARRDC3 | 4610390 | 4117 | 0.150 | -0.0360 | No | ||
| 25 | DDR2 | 7050273 | 4279 | 0.138 | -0.0426 | No | ||
| 26 | SOX10 | 6200538 | 4333 | 0.134 | -0.0435 | No | ||
| 27 | ZADH2 | 520438 | 4417 | 0.128 | -0.0460 | No | ||
| 28 | PDE4D | 2470528 6660014 | 4424 | 0.128 | -0.0444 | No | ||
| 29 | PRLR | 1980647 1990097 | 4485 | 0.124 | -0.0457 | No | ||
| 30 | COL4A3 | 5910075 | 4530 | 0.121 | -0.0463 | No | ||
| 31 | NFIL3 | 4070377 | 4541 | 0.120 | -0.0450 | No | ||
| 32 | PAFAH2 | 3140451 5720593 6510014 | 4820 | 0.104 | -0.0585 | No | ||
| 33 | PCF11 | 1240180 | 4841 | 0.103 | -0.0580 | No | ||
| 34 | PLA2G2E | 4060128 | 4866 | 0.102 | -0.0577 | No | ||
| 35 | COL4A4 | 1050541 | 4919 | 0.099 | -0.0590 | No | ||
| 36 | HNRPR | 2320440 2900601 4610008 | 5017 | 0.094 | -0.0629 | No | ||
| 37 | LEMD2 | 1940156 | 5039 | 0.093 | -0.0626 | No | ||
| 38 | TMPRSS4 | 1050402 1710128 2480341 | 5042 | 0.093 | -0.0613 | No | ||
| 39 | GYS1 | 540154 | 5274 | 0.084 | -0.0725 | No | ||
| 40 | PPP1CB | 3130411 3780270 4150180 | 5305 | 0.082 | -0.0729 | No | ||
| 41 | FGF14 | 2630390 3520075 6770048 | 5366 | 0.079 | -0.0749 | No | ||
| 42 | HOXC13 | 4070722 | 5476 | 0.076 | -0.0797 | No | ||
| 43 | ASGR1 | 1850088 | 5509 | 0.074 | -0.0803 | No | ||
| 44 | TAC1 | 7000195 380706 | 5645 | 0.069 | -0.0866 | No | ||
| 45 | BDNF | 2940128 3520368 | 5794 | 0.064 | -0.0936 | No | ||
| 46 | TRPM1 | 50746 2060301 5130100 5720129 | 5872 | 0.062 | -0.0968 | No | ||
| 47 | CASK | 2340215 3290576 6770215 | 5887 | 0.061 | -0.0967 | No | ||
| 48 | CDKL5 | 2450070 | 5912 | 0.060 | -0.0970 | No | ||
| 49 | IL19 | 6040142 | 5944 | 0.059 | -0.0978 | No | ||
| 50 | PITX2 | 870537 2690139 | 6007 | 0.058 | -0.1003 | No | ||
| 51 | EDN2 | 6760647 | 6102 | 0.055 | -0.1046 | No | ||
| 52 | C1QL1 | 2030465 | 6433 | 0.047 | -0.1217 | No | ||
| 53 | DBH | 3610458 | 6448 | 0.047 | -0.1218 | No | ||
| 54 | SULF1 | 430575 | 6526 | 0.045 | -0.1253 | No | ||
| 55 | PFN2 | 730040 5700026 | 6529 | 0.045 | -0.1247 | No | ||
| 56 | CBX4 | 70332 | 6634 | 0.043 | -0.1297 | No | ||
| 57 | ARNT | 1170672 5670711 | 6708 | 0.041 | -0.1330 | No | ||
| 58 | UBQLN1 | 110280 730376 2470133 6590594 | 6893 | 0.037 | -0.1424 | No | ||
| 59 | MRC2 | 110369 2480520 | 6913 | 0.037 | -0.1429 | No | ||
| 60 | SP8 | 4060576 | 7014 | 0.035 | -0.1478 | No | ||
| 61 | SLC12A1 | 3450017 6620133 6770148 | 7032 | 0.035 | -0.1482 | No | ||
| 62 | ALDH1A2 | 2320301 | 7064 | 0.034 | -0.1493 | No | ||
| 63 | MOSPD2 | 4010064 | 7208 | 0.031 | -0.1566 | No | ||
| 64 | CHRM1 | 4280619 | 7248 | 0.030 | -0.1583 | No | ||
| 65 | SERPINA7 | 2510537 | 7287 | 0.029 | -0.1599 | No | ||
| 66 | RFX4 | 3710100 5340215 | 7342 | 0.028 | -0.1624 | No | ||
| 67 | HOXA5 | 6840026 | 7482 | 0.026 | -0.1695 | No | ||
| 68 | FGB | 7000010 | 7488 | 0.026 | -0.1694 | No | ||
| 69 | KCNJ2 | 630019 | 7545 | 0.025 | -0.1721 | No | ||
| 70 | MEOX2 | 6400288 | 7561 | 0.025 | -0.1725 | No | ||
| 71 | RAD23B | 2190671 | 7690 | 0.023 | -0.1791 | No | ||
| 72 | PLA2G4A | 6380364 | 7770 | 0.021 | -0.1830 | No | ||
| 73 | PHOX2B | 5270075 | 8109 | 0.016 | -0.2011 | No | ||
| 74 | ELAVL2 | 360181 | 8471 | 0.011 | -0.2205 | No | ||
| 75 | EHF | 4560088 | 8489 | 0.011 | -0.2213 | No | ||
| 76 | PDCL | 2340348 | 8581 | 0.009 | -0.2261 | No | ||
| 77 | IL1RAPL2 | 1940075 | 8738 | 0.007 | -0.2345 | No | ||
| 78 | NHLH1 | 6100452 | 8876 | 0.005 | -0.2418 | No | ||
| 79 | ANGPT1 | 3990368 5130288 6770035 | 8885 | 0.005 | -0.2422 | No | ||
| 80 | SLC19A3 | 2650601 | 9171 | 0.001 | -0.2576 | No | ||
| 81 | SDPR | 3360292 | 9867 | -0.008 | -0.2952 | No | ||
| 82 | PHF16 | 50373 7200161 | 9999 | -0.010 | -0.3021 | No | ||
| 83 | FCER1G | 5550020 | 10022 | -0.011 | -0.3032 | No | ||
| 84 | BDKRB1 | 730592 | 10051 | -0.011 | -0.3045 | No | ||
| 85 | P2RY4 | 630053 | 10238 | -0.014 | -0.3144 | No | ||
| 86 | CYP24A1 | 2340338 | 10493 | -0.018 | -0.3279 | No | ||
| 87 | RGR | 730156 6400368 | 10561 | -0.019 | -0.3313 | No | ||
| 88 | IL27 | 1990324 | 10643 | -0.020 | -0.3353 | No | ||
| 89 | CP | 2570484 | 11373 | -0.031 | -0.3744 | No | ||
| 90 | SMAD1 | 630537 1850333 | 11611 | -0.036 | -0.3867 | No | ||
| 91 | CHD2 | 4070039 | 11727 | -0.038 | -0.3924 | No | ||
| 92 | PPL | 2360072 | 11777 | -0.039 | -0.3944 | No | ||
| 93 | LEP | 4010053 | 11801 | -0.040 | -0.3951 | No | ||
| 94 | SERPINF1 | 7040367 | 11826 | -0.040 | -0.3958 | No | ||
| 95 | PIP5K1A | 4560672 | 11935 | -0.042 | -0.4010 | No | ||
| 96 | KCNH7 | 7510068 | 12172 | -0.047 | -0.4130 | No | ||
| 97 | TRIB1 | 2320435 | 12272 | -0.050 | -0.4176 | No | ||
| 98 | ASAH2 | 3130452 | 12580 | -0.057 | -0.4334 | No | ||
| 99 | SMARCA1 | 4230315 | 12587 | -0.058 | -0.4329 | No | ||
| 100 | KCNE3 | 5550195 | 12643 | -0.059 | -0.4349 | No | ||
| 101 | ESR1 | 4060372 5860193 | 12674 | -0.060 | -0.4357 | No | ||
| 102 | RBPMS | 3990494 | 12692 | -0.061 | -0.4357 | No | ||
| 103 | DMD | 1740041 3990332 | 12741 | -0.062 | -0.4373 | No | ||
| 104 | LMO4 | 3800746 | 12781 | -0.064 | -0.4385 | No | ||
| 105 | CITED1 | 2350670 | 12797 | -0.064 | -0.4383 | No | ||
| 106 | SPRY4 | 1570594 | 12895 | -0.067 | -0.4425 | No | ||
| 107 | ITGA11 | 1740112 6590110 | 12907 | -0.067 | -0.4421 | No | ||
| 108 | FIGN | 7000601 | 12927 | -0.068 | -0.4421 | No | ||
| 109 | MBNL1 | 2640762 7100048 | 13161 | -0.075 | -0.4536 | No | ||
| 110 | SMAD6 | 870504 | 13179 | -0.076 | -0.4533 | No | ||
| 111 | PKNOX2 | 610066 2810075 4230601 | 13212 | -0.077 | -0.4539 | No | ||
| 112 | BNC2 | 4810603 | 13271 | -0.079 | -0.4558 | No | ||
| 113 | GLP2R | 4150577 5290273 7050095 | 13533 | -0.090 | -0.4686 | No | ||
| 114 | SOX5 | 2370576 2900167 3190128 5050528 | 13552 | -0.091 | -0.4682 | No | ||
| 115 | CSMD3 | 1660427 1940687 | 13555 | -0.091 | -0.4669 | No | ||
| 116 | AQP9 | 2650047 | 13709 | -0.097 | -0.4738 | No | ||
| 117 | ONECUT2 | 1340008 | 13769 | -0.100 | -0.4754 | No | ||
| 118 | PCTP | 6980324 | 13834 | -0.103 | -0.4773 | No | ||
| 119 | HAVCR1 | 5080497 | 13857 | -0.104 | -0.4769 | No | ||
| 120 | PDGFRL | 870168 4590128 | 13976 | -0.111 | -0.4816 | No | ||
| 121 | FST | 1110600 | 14043 | -0.116 | -0.4834 | No | ||
| 122 | PTPN12 | 2030309 6020725 6130746 6290609 | 14312 | -0.133 | -0.4959 | No | ||
| 123 | ASCL2 | 1400273 | 14512 | -0.148 | -0.5044 | No | ||
| 124 | VNN3 | 730053 4610609 | 14721 | -0.167 | -0.5132 | No | ||
| 125 | SLC6A4 | 2230372 | 14748 | -0.169 | -0.5120 | No | ||
| 126 | FMO2 | 3850338 | 14833 | -0.177 | -0.5138 | Yes | ||
| 127 | SYNCRIP | 1690195 3140113 4670279 | 14861 | -0.182 | -0.5125 | Yes | ||
| 128 | PCDH7 | 450398 4150040 | 14882 | -0.185 | -0.5108 | Yes | ||
| 129 | FGA | 4210056 5220239 | 14963 | -0.193 | -0.5122 | Yes | ||
| 130 | PDGFRA | 2940332 | 14994 | -0.197 | -0.5108 | Yes | ||
| 131 | PDGFB | 3060440 6370008 | 15042 | -0.203 | -0.5102 | Yes | ||
| 132 | HIST1H1C | 3870603 | 15060 | -0.204 | -0.5080 | Yes | ||
| 133 | TBR1 | 5130053 | 15068 | -0.205 | -0.5053 | Yes | ||
| 134 | MAP3K3 | 610685 | 15075 | -0.206 | -0.5025 | Yes | ||
| 135 | TOP1 | 770471 4060632 6650324 | 15099 | -0.209 | -0.5005 | Yes | ||
| 136 | EFNA5 | 460707 2190519 | 15207 | -0.225 | -0.5029 | Yes | ||
| 137 | SPRED1 | 6940706 | 15259 | -0.231 | -0.5021 | Yes | ||
| 138 | CACNB2 | 1500095 7330707 | 15276 | -0.234 | -0.4994 | Yes | ||
| 139 | NEK6 | 3360687 | 15498 | -0.272 | -0.5072 | Yes | ||
| 140 | PPARG | 1990168 2680603 6130632 | 15518 | -0.276 | -0.5040 | Yes | ||
| 141 | MTF1 | 520427 2340364 | 15546 | -0.280 | -0.5012 | Yes | ||
| 142 | RPS21 | 3120673 | 15593 | -0.288 | -0.4993 | Yes | ||
| 143 | ATAD2 | 870242 2260687 | 15795 | -0.334 | -0.5051 | Yes | ||
| 144 | UGP2 | 7000647 | 16013 | -0.386 | -0.5110 | Yes | ||
| 145 | STX18 | 4760706 | 16061 | -0.397 | -0.5074 | Yes | ||
| 146 | LUZP1 | 940075 4280458 6180070 | 16090 | -0.403 | -0.5028 | Yes | ||
| 147 | THRA | 6770341 110164 1990600 | 16183 | -0.427 | -0.5012 | Yes | ||
| 148 | TCF12 | 3610324 7000156 | 16359 | -0.484 | -0.5033 | Yes | ||
| 149 | BNIP3L | 1940347 | 16432 | -0.503 | -0.4995 | Yes | ||
| 150 | NFE2L2 | 2810619 3390162 | 16569 | -0.552 | -0.4985 | Yes | ||
| 151 | KPNA3 | 7040088 | 16574 | -0.553 | -0.4902 | Yes | ||
| 152 | MPP6 | 6020148 | 16618 | -0.568 | -0.4839 | Yes | ||
| 153 | MAP4K4 | 2360059 | 16829 | -0.647 | -0.4854 | Yes | ||
| 154 | BCL6 | 940100 | 16836 | -0.649 | -0.4758 | Yes | ||
| 155 | PRDX3 | 1690035 | 16991 | -0.717 | -0.4731 | Yes | ||
| 156 | RUVBL2 | 1190377 | 17097 | -0.768 | -0.4671 | Yes | ||
| 157 | MAP2K6 | 1230056 2940204 | 17105 | -0.770 | -0.4557 | Yes | ||
| 158 | CCL3 | 2810092 | 17168 | -0.800 | -0.4468 | Yes | ||
| 159 | H2AFZ | 1470168 | 17329 | -0.879 | -0.4421 | Yes | ||
| 160 | DYRK3 | 1090014 | 17436 | -0.947 | -0.4333 | Yes | ||
| 161 | S100A9 | 7050528 | 17444 | -0.953 | -0.4191 | Yes | ||
| 162 | RHOB | 1500309 | 17468 | -0.964 | -0.4056 | Yes | ||
| 163 | PPM1B | 380180 450154 3610215 | 17496 | -0.977 | -0.3922 | Yes | ||
| 164 | EIF4A2 | 1170494 1740711 2850504 | 17540 | -0.997 | -0.3792 | Yes | ||
| 165 | SLC7A11 | 2850138 | 17567 | -1.000 | -0.3654 | Yes | ||
| 166 | SLIT3 | 7100132 | 17604 | -1.015 | -0.3518 | Yes | ||
| 167 | SOCS2 | 4760692 | 17628 | -1.024 | -0.3374 | Yes | ||
| 168 | TOB1 | 4150138 | 17679 | -1.057 | -0.3239 | Yes | ||
| 169 | RPA3 | 5700136 | 17753 | -1.109 | -0.3109 | Yes | ||
| 170 | ARF6 | 3520026 | 17773 | -1.123 | -0.2948 | Yes | ||
| 171 | ACLY | 1740609 | 17817 | -1.163 | -0.2793 | Yes | ||
| 172 | PDAP1 | 1410735 | 17828 | -1.169 | -0.2620 | Yes | ||
| 173 | CSNK1E | 2850347 5050093 6110301 | 17859 | -1.190 | -0.2454 | Yes | ||
| 174 | CCND2 | 5340167 | 17917 | -1.231 | -0.2297 | Yes | ||
| 175 | UBE2E2 | 610053 | 18020 | -1.314 | -0.2151 | Yes | ||
| 176 | NFKBIA | 1570152 | 18024 | -1.317 | -0.1951 | Yes | ||
| 177 | SERTAD4 | 130593 | 18075 | -1.373 | -0.1768 | Yes | ||
| 178 | EIF4A1 | 1990341 2810300 | 18104 | -1.391 | -0.1571 | Yes | ||
| 179 | G0S2 | 6550020 | 18203 | -1.520 | -0.1392 | Yes | ||
| 180 | FOXP1 | 6510433 | 18275 | -1.604 | -0.1185 | Yes | ||
| 181 | RRBP1 | 2260477 5050440 | 18459 | -2.041 | -0.0972 | Yes | ||
| 182 | DUSP1 | 6860121 | 18491 | -2.179 | -0.0656 | Yes | ||
| 183 | H3F3B | 1410300 | 18538 | -2.349 | -0.0321 | Yes | ||
| 184 | PER1 | 3290315 | 18540 | -2.373 | 0.0041 | Yes |

