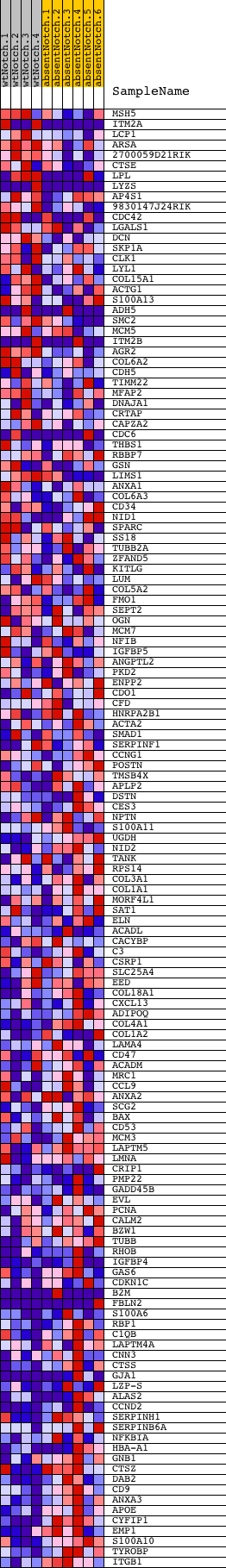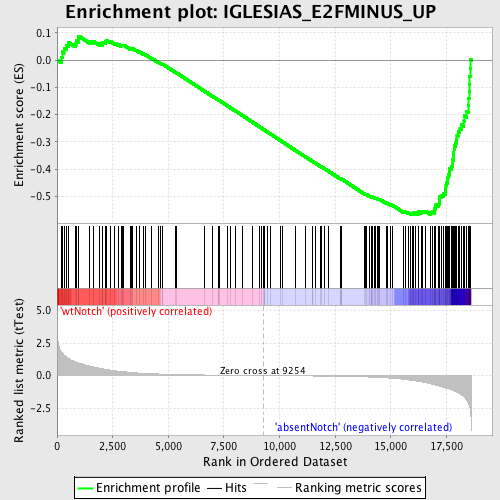
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #wtNotch_versus_absentNotch |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#wtNotch_versus_absentNotch |
| Upregulated in class | absentNotch |
| GeneSet | IGLESIAS_E2FMINUS_UP |
| Enrichment Score (ES) | -0.5663753 |
| Normalized Enrichment Score (NES) | -1.6649283 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.15132578 |
| FWER p-Value | 0.829 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MSH5 | 1340019 4730333 | 193 | 1.810 | 0.0110 | No | ||
| 2 | ITM2A | 460576 4210086 | 226 | 1.737 | 0.0298 | No | ||
| 3 | LCP1 | 4280397 | 310 | 1.578 | 0.0440 | No | ||
| 4 | ARSA | 5570717 | 439 | 1.377 | 0.0534 | No | ||
| 5 | 2700059D21RIK | 2260441 | 509 | 1.305 | 0.0651 | No | ||
| 6 | CTSE | 2030292 | 820 | 1.051 | 0.0608 | No | ||
| 7 | LPL | 1450735 2970706 | 856 | 1.023 | 0.0710 | No | ||
| 8 | LYZS | 50068 6770717 | 954 | 0.981 | 0.0774 | No | ||
| 9 | AP4S1 | 4730292 | 972 | 0.973 | 0.0880 | No | ||
| 10 | 9830147J24RIK | 2690451 | 1477 | 0.724 | 0.0693 | No | ||
| 11 | CDC42 | 1240168 3440278 4480519 5290162 | 1613 | 0.666 | 0.0698 | No | ||
| 12 | LGALS1 | 6450193 | 1921 | 0.558 | 0.0598 | No | ||
| 13 | DCN | 510332 5340026 5900711 6550092 | 2029 | 0.523 | 0.0602 | No | ||
| 14 | SKP1A | 2450102 | 2049 | 0.516 | 0.0653 | No | ||
| 15 | CLK1 | 1780551 2100102 2340347 2480347 | 2164 | 0.482 | 0.0649 | No | ||
| 16 | LYL1 | 2900242 | 2186 | 0.476 | 0.0694 | No | ||
| 17 | COL15A1 | 6840113 | 2210 | 0.468 | 0.0737 | No | ||
| 18 | ACTG1 | 540215 | 2384 | 0.417 | 0.0692 | No | ||
| 19 | S100A13 | 520019 | 2590 | 0.366 | 0.0625 | No | ||
| 20 | ADH5 | 6290100 6380711 | 2757 | 0.333 | 0.0574 | No | ||
| 21 | SMC2 | 4810133 | 2873 | 0.310 | 0.0549 | No | ||
| 22 | MCM5 | 2680647 | 2931 | 0.300 | 0.0553 | No | ||
| 23 | ITM2B | 1850482 5860671 6200014 | 2997 | 0.288 | 0.0552 | No | ||
| 24 | AGR2 | 3990161 | 3295 | 0.239 | 0.0420 | No | ||
| 25 | COL6A2 | 1780142 | 3334 | 0.234 | 0.0427 | No | ||
| 26 | CDH5 | 5340487 | 3396 | 0.225 | 0.0421 | No | ||
| 27 | TIMM22 | 1240397 | 3546 | 0.207 | 0.0364 | No | ||
| 28 | MFAP2 | 2970685 3060300 | 3686 | 0.191 | 0.0312 | No | ||
| 29 | DNAJA1 | 1660010 2320358 6840136 | 3871 | 0.173 | 0.0233 | No | ||
| 30 | CRTAP | 3990725 | 3968 | 0.165 | 0.0200 | No | ||
| 31 | CAPZA2 | 110427 4590064 | 4246 | 0.140 | 0.0067 | No | ||
| 32 | CDC6 | 4570296 5360600 | 4536 | 0.120 | -0.0075 | No | ||
| 33 | THBS1 | 4560494 430288 | 4657 | 0.114 | -0.0127 | No | ||
| 34 | RBBP7 | 430113 450450 2370309 | 4730 | 0.110 | -0.0153 | No | ||
| 35 | GSN | 3830168 | 4751 | 0.108 | -0.0151 | No | ||
| 36 | LIMS1 | 110138 1450398 | 5341 | 0.081 | -0.0460 | No | ||
| 37 | ANXA1 | 2320053 | 5372 | 0.079 | -0.0467 | No | ||
| 38 | COL6A3 | 2640717 4070064 5390717 | 6627 | 0.043 | -0.1141 | No | ||
| 39 | CD34 | 6650270 | 6970 | 0.036 | -0.1321 | No | ||
| 40 | NID1 | 60504 | 7250 | 0.030 | -0.1469 | No | ||
| 41 | SPARC | 1690086 | 7266 | 0.030 | -0.1474 | No | ||
| 42 | SS18 | 1940440 2900072 6900129 | 7307 | 0.029 | -0.1492 | No | ||
| 43 | TUBB2A | 6940435 | 7664 | 0.023 | -0.1682 | No | ||
| 44 | ZFAND5 | 6770673 | 7807 | 0.021 | -0.1756 | No | ||
| 45 | KITLG | 2120047 6220300 | 8023 | 0.018 | -0.1870 | No | ||
| 46 | LUM | 5420079 | 8343 | 0.013 | -0.2041 | No | ||
| 47 | COL5A2 | 780040 | 8775 | 0.006 | -0.2274 | No | ||
| 48 | FMO1 | 5360086 | 9090 | 0.002 | -0.2444 | No | ||
| 49 | SEPT2 | 2650152 3520142 | 9172 | 0.001 | -0.2487 | No | ||
| 50 | OGN | 3710326 3800193 | 9268 | -0.000 | -0.2539 | No | ||
| 51 | MCM7 | 3290292 5220056 | 9312 | -0.001 | -0.2562 | No | ||
| 52 | NFIB | 460450 | 9326 | -0.001 | -0.2569 | No | ||
| 53 | IGFBP5 | 2360592 | 9436 | -0.003 | -0.2628 | No | ||
| 54 | ANGPTL2 | 6980152 6760162 | 9591 | -0.005 | -0.2710 | No | ||
| 55 | PKD2 | 4670390 | 10035 | -0.011 | -0.2949 | No | ||
| 56 | ENPP2 | 5860546 | 10130 | -0.012 | -0.2998 | No | ||
| 57 | CDO1 | 2480279 | 10692 | -0.021 | -0.3300 | No | ||
| 58 | CFD | 2320736 | 11161 | -0.028 | -0.3550 | No | ||
| 59 | HNRPA2B1 | 1240619 6200161 6200494 6980408 | 11467 | -0.033 | -0.3711 | No | ||
| 60 | ACTA2 | 60008 1230082 | 11500 | -0.034 | -0.3724 | No | ||
| 61 | SMAD1 | 630537 1850333 | 11611 | -0.036 | -0.3779 | No | ||
| 62 | SERPINF1 | 7040367 | 11826 | -0.040 | -0.3890 | No | ||
| 63 | CCNG1 | 870402 1500685 6110594 | 11862 | -0.041 | -0.3904 | No | ||
| 64 | POSTN | 450411 6040451 | 12021 | -0.044 | -0.3985 | No | ||
| 65 | TMSB4X | 6620114 | 12177 | -0.048 | -0.4063 | No | ||
| 66 | APLP2 | 3610037 | 12715 | -0.061 | -0.4346 | No | ||
| 67 | DSTN | 3140592 | 12732 | -0.062 | -0.4348 | No | ||
| 68 | CES3 | 5050167 3290458 | 12770 | -0.063 | -0.4360 | No | ||
| 69 | NPTN | 6200373 | 12794 | -0.064 | -0.4365 | No | ||
| 70 | S100A11 | 2260064 | 13805 | -0.102 | -0.4900 | No | ||
| 71 | UGDH | 4730400 | 13858 | -0.105 | -0.4915 | No | ||
| 72 | NID2 | 2320059 | 13913 | -0.108 | -0.4932 | No | ||
| 73 | TANK | 1090020 2060458 2370400 | 14051 | -0.116 | -0.4992 | No | ||
| 74 | RPS14 | 430541 | 14139 | -0.122 | -0.5025 | No | ||
| 75 | COL3A1 | 6420273 6620044 | 14149 | -0.122 | -0.5015 | No | ||
| 76 | COL1A1 | 730020 | 14172 | -0.124 | -0.5013 | No | ||
| 77 | MORF4L1 | 3190100 | 14285 | -0.132 | -0.5058 | No | ||
| 78 | SAT1 | 4570463 | 14329 | -0.134 | -0.5065 | No | ||
| 79 | ELN | 5080347 | 14378 | -0.138 | -0.5075 | No | ||
| 80 | ACADL | 6420044 6860025 | 14458 | -0.144 | -0.5100 | No | ||
| 81 | CACYBP | 1410014 | 14493 | -0.147 | -0.5101 | No | ||
| 82 | C3 | 1740372 | 14801 | -0.174 | -0.5247 | No | ||
| 83 | CSRP1 | 2810403 | 14832 | -0.177 | -0.5242 | No | ||
| 84 | SLC25A4 | 2360519 | 15006 | -0.198 | -0.5312 | No | ||
| 85 | EED | 2320373 | 15059 | -0.204 | -0.5316 | No | ||
| 86 | COL18A1 | 610301 4570338 | 15582 | -0.286 | -0.5565 | No | ||
| 87 | CXCL13 | 6290402 | 15658 | -0.302 | -0.5570 | No | ||
| 88 | ADIPOQ | 6590519 | 15772 | -0.328 | -0.5592 | No | ||
| 89 | COL4A1 | 1740575 | 15894 | -0.355 | -0.5616 | Yes | ||
| 90 | COL1A2 | 380364 | 15969 | -0.373 | -0.5612 | Yes | ||
| 91 | LAMA4 | 3170647 | 16021 | -0.387 | -0.5593 | Yes | ||
| 92 | CD47 | 1850603 | 16127 | -0.413 | -0.5601 | Yes | ||
| 93 | ACADM | 460112 3390113 | 16226 | -0.439 | -0.5602 | Yes | ||
| 94 | MRC1 | 730097 | 16256 | -0.449 | -0.5565 | Yes | ||
| 95 | CCL9 | 4610725 | 16363 | -0.485 | -0.5565 | Yes | ||
| 96 | ANXA2 | 3140402 | 16428 | -0.502 | -0.5540 | Yes | ||
| 97 | SCG2 | 5670438 130671 | 16545 | -0.544 | -0.5538 | Yes | ||
| 98 | BAX | 3830008 | 16778 | -0.628 | -0.5589 | Yes | ||
| 99 | CD53 | 5550162 | 16850 | -0.657 | -0.5550 | Yes | ||
| 100 | MCM3 | 5570068 | 16965 | -0.703 | -0.5528 | Yes | ||
| 101 | LAPTM5 | 5550372 | 16966 | -0.704 | -0.5445 | Yes | ||
| 102 | LMNA | 520471 1500075 3190167 4210020 | 17007 | -0.724 | -0.5381 | Yes | ||
| 103 | CRIP1 | 4590377 | 17028 | -0.736 | -0.5305 | Yes | ||
| 104 | PMP22 | 4010239 6550072 | 17164 | -0.797 | -0.5283 | Yes | ||
| 105 | GADD45B | 2350408 | 17173 | -0.803 | -0.5193 | Yes | ||
| 106 | EVL | 1740113 | 17197 | -0.818 | -0.5108 | Yes | ||
| 107 | PCNA | 940754 | 17203 | -0.821 | -0.5014 | Yes | ||
| 108 | CALM2 | 6620463 | 17272 | -0.852 | -0.4950 | Yes | ||
| 109 | BZW1 | 460270 | 17384 | -0.913 | -0.4902 | Yes | ||
| 110 | TUBB | 730670 3390037 | 17452 | -0.957 | -0.4825 | Yes | ||
| 111 | RHOB | 1500309 | 17468 | -0.964 | -0.4719 | Yes | ||
| 112 | IGFBP4 | 2680309 5420441 | 17478 | -0.968 | -0.4609 | Yes | ||
| 113 | GAS6 | 4480021 | 17500 | -0.979 | -0.4504 | Yes | ||
| 114 | CDKN1C | 6520577 | 17550 | -0.999 | -0.4413 | Yes | ||
| 115 | B2M | 5080332 5130059 | 17565 | -1.000 | -0.4302 | Yes | ||
| 116 | FBLN2 | 730601 3440215 | 17585 | -1.005 | -0.4193 | Yes | ||
| 117 | S100A6 | 1690204 | 17636 | -1.028 | -0.4098 | Yes | ||
| 118 | RBP1 | 1690072 | 17652 | -1.039 | -0.3983 | Yes | ||
| 119 | C1QB | 5910292 | 17725 | -1.091 | -0.3893 | Yes | ||
| 120 | LAPTM4A | 450092 | 17754 | -1.109 | -0.3777 | Yes | ||
| 121 | CNN3 | 6110020 | 17777 | -1.131 | -0.3655 | Yes | ||
| 122 | CTSS | 1740056 | 17805 | -1.154 | -0.3533 | Yes | ||
| 123 | GJA1 | 5220731 | 17813 | -1.160 | -0.3399 | Yes | ||
| 124 | LZP-S | 5700609 6020195 | 17839 | -1.181 | -0.3273 | Yes | ||
| 125 | ALAS2 | 6550176 | 17846 | -1.185 | -0.3136 | Yes | ||
| 126 | CCND2 | 5340167 | 17917 | -1.231 | -0.3028 | Yes | ||
| 127 | SERPINH1 | 6130014 | 17953 | -1.257 | -0.2898 | Yes | ||
| 128 | SERPINB6A | 1780500 | 17971 | -1.269 | -0.2757 | Yes | ||
| 129 | NFKBIA | 1570152 | 18024 | -1.317 | -0.2629 | Yes | ||
| 130 | HBA-A1 | 360435 | 18107 | -1.393 | -0.2509 | Yes | ||
| 131 | GNB1 | 2120397 | 18190 | -1.508 | -0.2375 | Yes | ||
| 132 | CTSZ | 1500687 1690364 | 18288 | -1.634 | -0.2234 | Yes | ||
| 133 | DAB2 | 60309 | 18304 | -1.664 | -0.2045 | Yes | ||
| 134 | CD9 | 4730041 | 18408 | -1.912 | -0.1874 | Yes | ||
| 135 | ANXA3 | 5570494 | 18481 | -2.144 | -0.1659 | Yes | ||
| 136 | APOE | 4200671 | 18507 | -2.229 | -0.1409 | Yes | ||
| 137 | CYFIP1 | 5690082 | 18521 | -2.260 | -0.1148 | Yes | ||
| 138 | EMP1 | 4120438 | 18529 | -2.294 | -0.0880 | Yes | ||
| 139 | S100A10 | 6350010 | 18545 | -2.418 | -0.0602 | Yes | ||
| 140 | TYROBP | 2230020 | 18566 | -2.584 | -0.0307 | Yes | ||
| 141 | ITGB1 | 5080156 6270528 | 18580 | -2.818 | 0.0019 | Yes |