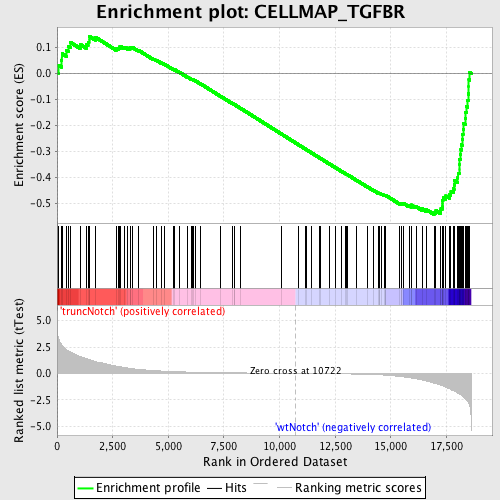
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #truncNotch_versus_wtNotch |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#truncNotch_versus_wtNotch |
| Upregulated in class | wtNotch |
| GeneSet | CELLMAP_TGFBR |
| Enrichment Score (ES) | -0.5438832 |
| Normalized Enrichment Score (NES) | -1.6260699 |
| Nominal p-value | 0.0028409092 |
| FDR q-value | 0.21169281 |
| FWER p-Value | 0.884 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ARRB2 | 2060441 | 48 | 3.434 | 0.0315 | No | ||
| 2 | CAMK2D | 2370685 | 198 | 2.728 | 0.0505 | No | ||
| 3 | ANAPC7 | 5260274 | 219 | 2.652 | 0.0758 | No | ||
| 4 | SMAD3 | 6450671 | 421 | 2.238 | 0.0871 | No | ||
| 5 | CDK4 | 540075 4540600 | 515 | 2.120 | 0.1031 | No | ||
| 6 | PIK3R2 | 1660193 | 593 | 2.025 | 0.1191 | No | ||
| 7 | SNIP1 | 5360092 | 1039 | 1.585 | 0.1108 | No | ||
| 8 | CCNE1 | 110064 | 1324 | 1.384 | 0.1092 | No | ||
| 9 | DAXX | 4010647 | 1391 | 1.342 | 0.1189 | No | ||
| 10 | STK11IP | 6980735 | 1434 | 1.321 | 0.1298 | No | ||
| 11 | FZR1 | 6220504 6660670 | 1436 | 1.320 | 0.1428 | No | ||
| 12 | FKBP1A | 2450368 | 1722 | 1.130 | 0.1386 | No | ||
| 13 | CAMK2G | 630097 | 2653 | 0.695 | 0.0953 | No | ||
| 14 | BTRC | 3170131 | 2756 | 0.646 | 0.0962 | No | ||
| 15 | VDR | 130156 510438 | 2783 | 0.637 | 0.1011 | No | ||
| 16 | TGFB1 | 1940162 | 2867 | 0.610 | 0.1027 | No | ||
| 17 | DAB2 | 60309 | 3033 | 0.551 | 0.0992 | No | ||
| 18 | JUNB | 4230048 | 3164 | 0.504 | 0.0972 | No | ||
| 19 | ETS1 | 5270278 6450717 6620465 | 3277 | 0.470 | 0.0958 | No | ||
| 20 | CDK6 | 4920253 | 3281 | 0.469 | 0.1003 | No | ||
| 21 | TP53 | 6130707 | 3380 | 0.442 | 0.0994 | No | ||
| 22 | FOS | 1850315 | 3645 | 0.386 | 0.0890 | No | ||
| 23 | MEF2A | 2350041 3360128 6040180 | 4341 | 0.266 | 0.0541 | No | ||
| 24 | ZEB2 | 4120070 4150215 | 4463 | 0.250 | 0.0500 | No | ||
| 25 | FOXO3 | 2510484 4480451 | 4697 | 0.222 | 0.0396 | No | ||
| 26 | HSPA8 | 2120315 1050132 6550746 | 4841 | 0.208 | 0.0340 | No | ||
| 27 | SMURF2 | 1940161 | 5211 | 0.172 | 0.0158 | No | ||
| 28 | SNX6 | 6200086 | 5290 | 0.166 | 0.0132 | No | ||
| 29 | SDC2 | 1980168 | 5487 | 0.151 | 0.0041 | No | ||
| 30 | CD44 | 3990072 4850671 5860411 6860148 7050551 | 5857 | 0.127 | -0.0146 | No | ||
| 31 | YAP1 | 4230577 | 6023 | 0.118 | -0.0223 | No | ||
| 32 | ANAPC2 | 6660438 | 6066 | 0.116 | -0.0234 | No | ||
| 33 | JUN | 840170 | 6110 | 0.114 | -0.0246 | No | ||
| 34 | ENG | 4280270 | 6200 | 0.109 | -0.0283 | No | ||
| 35 | ATF3 | 1940546 | 6450 | 0.097 | -0.0408 | No | ||
| 36 | SMAD6 | 870504 | 7349 | 0.066 | -0.0887 | No | ||
| 37 | PRKAR1B | 6130411 | 7881 | 0.052 | -0.1169 | No | ||
| 38 | CAV1 | 870025 | 7984 | 0.050 | -0.1219 | No | ||
| 39 | CITED1 | 2350670 | 7992 | 0.050 | -0.1218 | No | ||
| 40 | ZFYVE9 | 6980288 7100017 | 8253 | 0.043 | -0.1354 | No | ||
| 41 | ESR1 | 4060372 5860193 | 10104 | 0.010 | -0.2352 | No | ||
| 42 | XPO1 | 540707 | 10863 | -0.002 | -0.2762 | No | ||
| 43 | UBE2D1 | 1850400 | 11164 | -0.007 | -0.2923 | No | ||
| 44 | SMAD7 | 430377 | 11203 | -0.008 | -0.2943 | No | ||
| 45 | SKIL | 1090364 | 11420 | -0.012 | -0.3058 | No | ||
| 46 | CDKN1A | 4050088 6400706 | 11801 | -0.019 | -0.3262 | No | ||
| 47 | ANAPC10 | 870086 1170037 2260129 | 11843 | -0.019 | -0.3282 | No | ||
| 48 | MYC | 380541 4670170 | 12234 | -0.028 | -0.3490 | No | ||
| 49 | NUP153 | 7000452 | 12531 | -0.035 | -0.3646 | No | ||
| 50 | TGFBR3 | 5290577 | 12773 | -0.041 | -0.3772 | No | ||
| 51 | NFYC | 1400746 | 12956 | -0.047 | -0.3866 | No | ||
| 52 | FOSB | 1940142 | 13018 | -0.049 | -0.3894 | No | ||
| 53 | CCNB2 | 6510528 | 13046 | -0.050 | -0.3904 | No | ||
| 54 | CDC23 | 3190593 | 13459 | -0.066 | -0.4120 | No | ||
| 55 | EID2 | 3130176 | 13968 | -0.092 | -0.4385 | No | ||
| 56 | TGFBR1 | 1400148 4280020 6550711 | 14213 | -0.109 | -0.4506 | No | ||
| 57 | RBL2 | 580446 1400670 | 14429 | -0.128 | -0.4610 | No | ||
| 58 | RBX1 | 2120010 2340047 | 14472 | -0.132 | -0.4619 | No | ||
| 59 | HOXA9 | 610494 4730040 5130601 | 14495 | -0.134 | -0.4618 | No | ||
| 60 | MAP2K6 | 1230056 2940204 | 14593 | -0.145 | -0.4656 | No | ||
| 61 | TGFBR2 | 1780711 1980537 6550398 | 14702 | -0.157 | -0.4698 | No | ||
| 62 | RUNX2 | 1400193 4230215 | 14743 | -0.162 | -0.4704 | No | ||
| 63 | SNX1 | 3190670 | 14779 | -0.169 | -0.4706 | No | ||
| 64 | CDK2 | 130484 2260301 4010088 5050110 | 15400 | -0.284 | -0.5013 | No | ||
| 65 | AXIN1 | 3360358 6940451 | 15468 | -0.299 | -0.5019 | No | ||
| 66 | CAMK2B | 2760041 | 15490 | -0.306 | -0.5000 | No | ||
| 67 | AR | 6380167 | 15557 | -0.323 | -0.5004 | No | ||
| 68 | ATF2 | 2260348 2480441 | 15819 | -0.409 | -0.5104 | No | ||
| 69 | TGFB3 | 1070041 | 15914 | -0.438 | -0.5112 | No | ||
| 70 | SNW1 | 4010736 | 15922 | -0.442 | -0.5072 | No | ||
| 71 | PRKAR2A | 2340136 | 16133 | -0.512 | -0.5134 | No | ||
| 72 | CDC25A | 3800184 | 16404 | -0.617 | -0.5219 | No | ||
| 73 | MAPK8 | 2640195 | 16603 | -0.718 | -0.5255 | No | ||
| 74 | TFDP1 | 1980112 | 16945 | -0.926 | -0.5347 | Yes | ||
| 75 | CDC16 | 1940706 | 17027 | -0.980 | -0.5293 | Yes | ||
| 76 | CREBBP | 5690035 7040050 | 17238 | -1.091 | -0.5299 | Yes | ||
| 77 | UBE2D2 | 4590671 6510446 | 17253 | -1.102 | -0.5197 | Yes | ||
| 78 | MAPK14 | 5290731 | 17315 | -1.150 | -0.5116 | Yes | ||
| 79 | MAP3K7IP1 | 2120095 5890452 6040167 | 17322 | -1.155 | -0.5004 | Yes | ||
| 80 | SP1 | 6590017 | 17325 | -1.160 | -0.4890 | Yes | ||
| 81 | UBE2E1 | 4230064 1090440 | 17347 | -1.186 | -0.4784 | Yes | ||
| 82 | NFYA | 5860368 | 17477 | -1.293 | -0.4725 | Yes | ||
| 83 | HDAC1 | 2850670 | 17640 | -1.456 | -0.4668 | Yes | ||
| 84 | PRKCD | 770592 | 17702 | -1.525 | -0.4550 | Yes | ||
| 85 | SMAD2 | 4200592 | 17810 | -1.643 | -0.4445 | Yes | ||
| 86 | CUL1 | 1990632 | 17844 | -1.677 | -0.4296 | Yes | ||
| 87 | COPS5 | 1340722 5050215 | 17865 | -1.699 | -0.4138 | Yes | ||
| 88 | PIAS1 | 5340301 | 18009 | -1.854 | -0.4031 | Yes | ||
| 89 | ANAPC5 | 730164 7100685 | 18019 | -1.862 | -0.3851 | Yes | ||
| 90 | NCOA1 | 3610438 | 18073 | -1.911 | -0.3690 | Yes | ||
| 91 | UBE2D3 | 3190452 | 18074 | -1.915 | -0.3500 | Yes | ||
| 92 | MAP3K7 | 6040068 | 18082 | -1.929 | -0.3313 | Yes | ||
| 93 | UBE2L3 | 1690707 5900242 | 18125 | -2.003 | -0.3136 | Yes | ||
| 94 | SNX4 | 1980358 | 18147 | -2.037 | -0.2945 | Yes | ||
| 95 | SNX2 | 2470068 3360364 6510064 | 18172 | -2.069 | -0.2753 | Yes | ||
| 96 | FNTA | 70059 | 18207 | -2.120 | -0.2561 | Yes | ||
| 97 | RB1 | 5900338 | 18226 | -2.152 | -0.2357 | Yes | ||
| 98 | ZEB1 | 6840162 | 18249 | -2.192 | -0.2151 | Yes | ||
| 99 | CCND1 | 460524 770309 3120576 6980398 | 18270 | -2.224 | -0.1941 | Yes | ||
| 100 | ANAPC4 | 4540338 | 18344 | -2.384 | -0.1744 | Yes | ||
| 101 | STRAP | 1340170 | 18364 | -2.428 | -0.1513 | Yes | ||
| 102 | RBL1 | 3130372 | 18396 | -2.502 | -0.1282 | Yes | ||
| 103 | CTCF | 5340017 | 18444 | -2.618 | -0.1047 | Yes | ||
| 104 | NFYB | 1850053 | 18478 | -2.739 | -0.0793 | Yes | ||
| 105 | KPNB1 | 1690138 | 18506 | -2.828 | -0.0527 | Yes | ||
| 106 | ANAPC1 | 6350162 | 18510 | -2.840 | -0.0247 | Yes | ||
| 107 | SMAD4 | 5670519 | 18548 | -3.059 | 0.0037 | Yes |