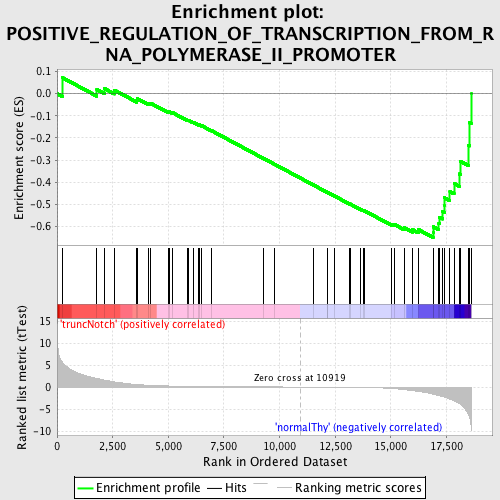
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
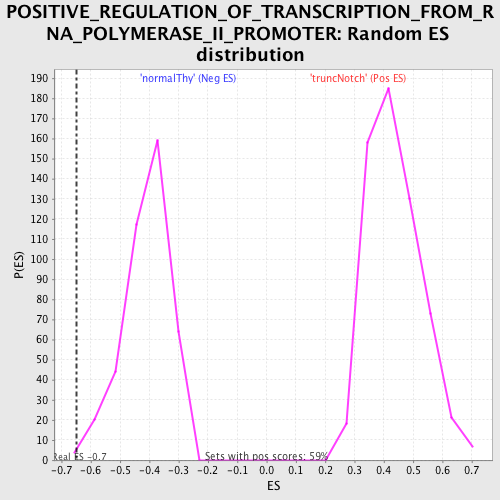
| GeneSet | POSITIVE_REGULATION_OF_TRANSCRIPTION_FROM_RNA_POLYMERASE_II_PROMOTER |
| Enrichment Score (ES) | -0.65005285 |
| Normalized Enrichment Score (NES) | -1.5899572 |
| Nominal p-value | 0.0024509805 |
| FDR q-value | 0.4803269 |
| FWER p-Value | 0.952 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TP53 | 6130707 | 231 | 5.694 | 0.0724 | No | ||
| 2 | ERCC2 | 2360750 4060390 6550138 | 1786 | 1.946 | 0.0178 | No | ||
| 3 | RBM14 | 5340731 5690315 | 2122 | 1.564 | 0.0230 | No | ||
| 4 | CCNH | 3190347 | 2583 | 1.142 | 0.0153 | No | ||
| 5 | EPAS1 | 5290156 | 3587 | 0.565 | -0.0303 | No | ||
| 6 | NCOA6 | 1780333 6450110 | 3611 | 0.552 | -0.0233 | No | ||
| 7 | GTF2H1 | 2570746 2650148 | 4099 | 0.405 | -0.0435 | No | ||
| 8 | NPAS2 | 770670 | 4215 | 0.377 | -0.0441 | No | ||
| 9 | IL4 | 6020537 | 4994 | 0.240 | -0.0824 | No | ||
| 10 | GLI2 | 3060632 | 5055 | 0.232 | -0.0822 | No | ||
| 11 | GLIS1 | 840471 | 5179 | 0.217 | -0.0856 | No | ||
| 12 | ELL3 | 1770484 | 5863 | 0.147 | -0.1202 | No | ||
| 13 | HNF4A | 6620440 | 5895 | 0.145 | -0.1197 | No | ||
| 14 | PLAGL1 | 3190082 6200193 | 6118 | 0.128 | -0.1297 | No | ||
| 15 | NUFIP1 | 6350241 | 6369 | 0.114 | -0.1415 | No | ||
| 16 | THRAP3 | 6180309 | 6378 | 0.114 | -0.1402 | No | ||
| 17 | UTF1 | 5700593 | 6500 | 0.108 | -0.1451 | No | ||
| 18 | CDK7 | 2640451 | 6945 | 0.088 | -0.1677 | No | ||
| 19 | MKL2 | 5700204 | 9262 | 0.028 | -0.2920 | No | ||
| 20 | HIF1A | 5670605 | 9785 | 0.019 | -0.3199 | No | ||
| 21 | BMP6 | 2100128 | 11502 | -0.010 | -0.4121 | No | ||
| 22 | CLOCK | 1410070 2940292 3390075 | 12131 | -0.023 | -0.4456 | No | ||
| 23 | FOXH1 | 2060180 | 12170 | -0.024 | -0.4473 | No | ||
| 24 | RXRA | 3800471 | 12488 | -0.032 | -0.4639 | No | ||
| 25 | GLI1 | 4560176 | 13136 | -0.053 | -0.4979 | No | ||
| 26 | FOXD3 | 6550156 | 13166 | -0.054 | -0.4987 | No | ||
| 27 | NRIP1 | 3190039 | 13633 | -0.079 | -0.5226 | No | ||
| 28 | PPARGC1B | 3290114 | 13778 | -0.090 | -0.5290 | No | ||
| 29 | FOXO3 | 2510484 4480451 | 13824 | -0.093 | -0.5301 | No | ||
| 30 | ELF4 | 4850193 | 15042 | -0.324 | -0.5908 | No | ||
| 31 | ATF4 | 730441 1740195 | 15175 | -0.374 | -0.5923 | No | ||
| 32 | ERCC3 | 6900008 | 15597 | -0.581 | -0.6063 | No | ||
| 33 | SQSTM1 | 6550056 | 15960 | -0.781 | -0.6142 | No | ||
| 34 | EPC1 | 1570050 5290095 6900193 | 16244 | -0.985 | -0.6147 | No | ||
| 35 | SMAD3 | 6450671 | 16901 | -1.640 | -0.6256 | Yes | ||
| 36 | MYO6 | 2190332 | 16909 | -1.647 | -0.6014 | Yes | ||
| 37 | SP1 | 6590017 | 17158 | -1.914 | -0.5863 | Yes | ||
| 38 | SUPT5H | 4150546 | 17187 | -1.943 | -0.5588 | Yes | ||
| 39 | ELF1 | 4780450 | 17335 | -2.163 | -0.5345 | Yes | ||
| 40 | SUPT4H1 | 6200056 | 17406 | -2.278 | -0.5043 | Yes | ||
| 41 | SMAD2 | 4200592 | 17412 | -2.286 | -0.4705 | Yes | ||
| 42 | MNAT1 | 730292 2350364 | 17656 | -2.745 | -0.4427 | Yes | ||
| 43 | MED6 | 6840500 | 17844 | -3.133 | -0.4060 | Yes | ||
| 44 | RUNX1 | 3840711 | 18089 | -3.768 | -0.3630 | Yes | ||
| 45 | MED12 | 4920095 | 18141 | -3.948 | -0.3069 | Yes | ||
| 46 | ARNTL | 3170463 | 18486 | -6.165 | -0.2336 | Yes | ||
| 47 | MAML1 | 2760008 | 18546 | -7.013 | -0.1322 | Yes | ||
| 48 | TNFRSF1A | 1090390 6520735 | 18612 | -9.119 | 0.0002 | Yes |