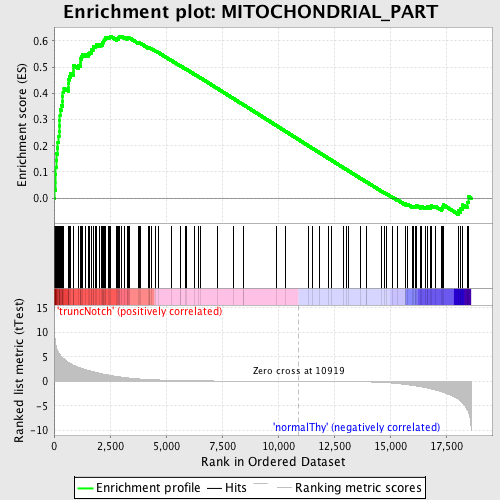
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | truncNotch |
| GeneSet | MITOCHONDRIAL_PART |
| Enrichment Score (ES) | 0.61843604 |
| Normalized Enrichment Score (NES) | 1.6388655 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.116554245 |
| FWER p-Value | 0.528 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | BAX | 3830008 | 36 | 8.812 | 0.0316 | Yes | ||
| 2 | TIMM50 | 1500139 | 53 | 8.098 | 0.0616 | Yes | ||
| 3 | ABCB6 | 6350138 | 55 | 8.001 | 0.0921 | Yes | ||
| 4 | MRPL55 | 2260075 | 80 | 7.257 | 0.1184 | Yes | ||
| 5 | PHB2 | 1230050 | 91 | 7.136 | 0.1451 | Yes | ||
| 6 | TIMM13 | 2450601 | 115 | 6.699 | 0.1693 | Yes | ||
| 7 | MRPL12 | 3120112 | 149 | 6.380 | 0.1919 | Yes | ||
| 8 | ATP5G2 | 840068 | 168 | 6.154 | 0.2144 | Yes | ||
| 9 | MRPS15 | 3840280 5360504 | 209 | 5.825 | 0.2344 | Yes | ||
| 10 | BCS1L | 5360102 | 221 | 5.735 | 0.2556 | Yes | ||
| 11 | ETFB | 2690025 | 247 | 5.606 | 0.2757 | Yes | ||
| 12 | ATP5G1 | 5290019 | 253 | 5.555 | 0.2966 | Yes | ||
| 13 | MRPS18A | 6770433 | 261 | 5.492 | 0.3171 | Yes | ||
| 14 | MRPS28 | 2230373 | 268 | 5.433 | 0.3375 | Yes | ||
| 15 | ATP5E | 4120411 | 333 | 4.999 | 0.3531 | Yes | ||
| 16 | UQCRC1 | 520053 | 374 | 4.792 | 0.3692 | Yes | ||
| 17 | SLC25A1 | 4050402 | 388 | 4.747 | 0.3865 | Yes | ||
| 18 | HTRA2 | 130048 1400025 | 395 | 4.702 | 0.4041 | Yes | ||
| 19 | SLC25A3 | 670112 | 439 | 4.578 | 0.4193 | Yes | ||
| 20 | TOMM22 | 1240142 | 638 | 3.890 | 0.4234 | Yes | ||
| 21 | ATP5D | 6550167 | 642 | 3.882 | 0.4380 | Yes | ||
| 22 | MRPL10 | 4120484 | 651 | 3.869 | 0.4523 | Yes | ||
| 23 | TIMM10 | 610070 1450019 | 680 | 3.769 | 0.4652 | Yes | ||
| 24 | NDUFS2 | 4850020 6200402 | 742 | 3.628 | 0.4757 | Yes | ||
| 25 | RAB11FIP5 | 4570600 | 856 | 3.315 | 0.4822 | Yes | ||
| 26 | NDUFV1 | 5420369 | 859 | 3.313 | 0.4947 | Yes | ||
| 27 | MRPS11 | 3130270 6660735 | 861 | 3.311 | 0.5073 | Yes | ||
| 28 | ABCF2 | 630040 3140035 6450601 | 1097 | 2.879 | 0.5055 | Yes | ||
| 29 | SURF1 | 5130048 | 1170 | 2.751 | 0.5121 | Yes | ||
| 30 | NDUFS3 | 1690136 5340528 | 1172 | 2.750 | 0.5226 | Yes | ||
| 31 | NDUFS8 | 4150100 | 1174 | 2.747 | 0.5330 | Yes | ||
| 32 | MRPS16 | 2510390 | 1235 | 2.641 | 0.5398 | Yes | ||
| 33 | VDAC2 | 7000215 | 1256 | 2.610 | 0.5487 | Yes | ||
| 34 | MRPS12 | 4210707 6110341 | 1412 | 2.383 | 0.5494 | Yes | ||
| 35 | ATP5O | 7000398 | 1544 | 2.218 | 0.5507 | Yes | ||
| 36 | MRPS35 | 2370603 4070551 | 1598 | 2.141 | 0.5560 | Yes | ||
| 37 | SDHD | 2570128 | 1664 | 2.059 | 0.5603 | Yes | ||
| 38 | BCKDHA | 50189 | 1670 | 2.054 | 0.5679 | Yes | ||
| 39 | MRPS24 | 5340022 | 1739 | 1.988 | 0.5718 | Yes | ||
| 40 | MRPS10 | 2060315 4880300 6040047 | 1767 | 1.961 | 0.5778 | Yes | ||
| 41 | PPOX | 2640678 3850102 | 1854 | 1.867 | 0.5803 | Yes | ||
| 42 | VDAC3 | 5390088 | 1896 | 1.818 | 0.5850 | Yes | ||
| 43 | NDUFS7 | 1740142 | 2018 | 1.698 | 0.5849 | Yes | ||
| 44 | SUPV3L1 | 1170072 | 2097 | 1.591 | 0.5867 | Yes | ||
| 45 | MRPL23 | 4070142 | 2151 | 1.534 | 0.5897 | Yes | ||
| 46 | NDUFA6 | 3990348 | 2161 | 1.527 | 0.5950 | Yes | ||
| 47 | ATP5J | 4210100 6900288 | 2216 | 1.484 | 0.5978 | Yes | ||
| 48 | DNAJA3 | 1980671 6220338 | 2226 | 1.478 | 0.6029 | Yes | ||
| 49 | TIMM17A | 130092 | 2262 | 1.446 | 0.6065 | Yes | ||
| 50 | ATP5A1 | 1990722 | 2302 | 1.410 | 0.6098 | Yes | ||
| 51 | BCKDK | 1940601 3990600 | 2306 | 1.409 | 0.6150 | Yes | ||
| 52 | SLC25A11 | 130097 770685 | 2424 | 1.283 | 0.6136 | Yes | ||
| 53 | COX15 | 2360402 | 2485 | 1.215 | 0.6150 | Yes | ||
| 54 | POLG2 | 5690047 | 2506 | 1.197 | 0.6184 | Yes | ||
| 55 | MFN2 | 2260195 6100164 | 2798 | 1.000 | 0.6065 | No | ||
| 56 | MRPL52 | 1170204 | 2818 | 0.993 | 0.6093 | No | ||
| 57 | CYCS | 2030072 | 2866 | 0.970 | 0.6104 | No | ||
| 58 | MRPL40 | 2370215 | 2885 | 0.957 | 0.6131 | No | ||
| 59 | ATP5F1 | 7000725 | 2904 | 0.946 | 0.6157 | No | ||
| 60 | ALDH4A1 | 2450450 | 2925 | 0.936 | 0.6182 | No | ||
| 61 | CS | 5080600 | 3021 | 0.868 | 0.6164 | No | ||
| 62 | ABCB7 | 4150324 | 3130 | 0.794 | 0.6136 | No | ||
| 63 | BCKDHB | 780372 | 3257 | 0.718 | 0.6095 | No | ||
| 64 | MRPS18C | 6760044 | 3283 | 0.709 | 0.6108 | No | ||
| 65 | ETFA | 50347 5690040 6370044 | 3309 | 0.696 | 0.6121 | No | ||
| 66 | TIMM17B | 6040400 | 3370 | 0.659 | 0.6114 | No | ||
| 67 | AIFM2 | 360537 3780520 4230138 6130082 | 3765 | 0.505 | 0.5920 | No | ||
| 68 | GRPEL1 | 3180240 | 3792 | 0.496 | 0.5925 | No | ||
| 69 | SLC25A15 | 4640368 5550064 | 3838 | 0.479 | 0.5919 | No | ||
| 70 | TIMM9 | 2450484 | 4219 | 0.376 | 0.5728 | No | ||
| 71 | ATP5C1 | 5910601 | 4241 | 0.371 | 0.5730 | No | ||
| 72 | TIMM23 | 60288 7040133 | 4251 | 0.367 | 0.5740 | No | ||
| 73 | MAOB | 2970520 3060056 | 4326 | 0.347 | 0.5713 | No | ||
| 74 | NDUFA9 | 2260280 | 4528 | 0.311 | 0.5616 | No | ||
| 75 | HSD3B1 | 3060672 | 4668 | 0.287 | 0.5552 | No | ||
| 76 | DBT | 5220364 5720440 | 5233 | 0.210 | 0.5255 | No | ||
| 77 | NR3C1 | 630563 | 5620 | 0.171 | 0.5052 | No | ||
| 78 | CENTA2 | 4920451 | 5636 | 0.169 | 0.5051 | No | ||
| 79 | HSD3B2 | 70068 770037 4120114 4230170 5690333 | 5850 | 0.148 | 0.4941 | No | ||
| 80 | PITRM1 | 3450400 | 5903 | 0.144 | 0.4918 | No | ||
| 81 | VDAC1 | 4760059 | 6263 | 0.120 | 0.4729 | No | ||
| 82 | UQCRB | 1450047 | 6430 | 0.112 | 0.4643 | No | ||
| 83 | ATP5G3 | 4230551 | 6523 | 0.107 | 0.4598 | No | ||
| 84 | UCP3 | 4200128 4210148 | 7303 | 0.074 | 0.4179 | No | ||
| 85 | MTX2 | 6660288 | 8025 | 0.054 | 0.3791 | No | ||
| 86 | HCCS | 670397 3170341 | 8437 | 0.044 | 0.3570 | No | ||
| 87 | MRPS22 | 5910458 | 8453 | 0.044 | 0.3564 | No | ||
| 88 | GATM | 380671 | 9936 | 0.016 | 0.2763 | No | ||
| 89 | BNIP3 | 3140270 | 10326 | 0.010 | 0.2553 | No | ||
| 90 | PHB | 2260739 | 11377 | -0.008 | 0.1985 | No | ||
| 91 | MRPS36 | 7050072 | 11550 | -0.011 | 0.1893 | No | ||
| 92 | CASQ1 | 6290035 7000707 | 11860 | -0.017 | 0.1726 | No | ||
| 93 | TFB2M | 3830161 4670059 4760184 | 12242 | -0.025 | 0.1521 | No | ||
| 94 | SDHA | 7000056 | 12362 | -0.029 | 0.1458 | No | ||
| 95 | NDUFS1 | 2650253 | 12928 | -0.045 | 0.1154 | No | ||
| 96 | NFS1 | 1170465 2680563 | 13055 | -0.049 | 0.1088 | No | ||
| 97 | OPA1 | 2030433 3520390 4730537 | 13121 | -0.052 | 0.1055 | No | ||
| 98 | MRPL32 | 110041 | 13682 | -0.083 | 0.0755 | No | ||
| 99 | HADHB | 60064 2690670 | 13927 | -0.101 | 0.0627 | No | ||
| 100 | COX6B2 | 1340010 4120039 | 13952 | -0.104 | 0.0618 | No | ||
| 101 | NDUFAB1 | 1980091 | 14627 | -0.205 | 0.0261 | No | ||
| 102 | NNT | 540253 1170471 5550092 6760397 | 14727 | -0.231 | 0.0216 | No | ||
| 103 | OGDH | 3840333 6350100 | 14835 | -0.265 | 0.0168 | No | ||
| 104 | TOMM34 | 5690102 | 15126 | -0.354 | 0.0025 | No | ||
| 105 | PIN4 | 2370324 | 15315 | -0.431 | -0.0060 | No | ||
| 106 | SLC25A22 | 2360050 | 15689 | -0.632 | -0.0238 | No | ||
| 107 | TIMM8B | 1190364 | 15698 | -0.639 | -0.0218 | No | ||
| 108 | NDUFA1 | 6200411 | 15796 | -0.687 | -0.0244 | No | ||
| 109 | ACN9 | 6660180 | 15980 | -0.794 | -0.0313 | No | ||
| 110 | ATP5B | 3870138 | 16029 | -0.841 | -0.0307 | No | ||
| 111 | FIS1 | 2760017 4070286 | 16145 | -0.929 | -0.0334 | No | ||
| 112 | BCL2 | 730132 1570736 2470138 3800044 4810037 5690068 5860504 6650164 | 16152 | -0.932 | -0.0301 | No | ||
| 113 | UQCRH | 4210463 | 16167 | -0.943 | -0.0273 | No | ||
| 114 | RHOT2 | 1660053 2120292 | 16374 | -1.067 | -0.0344 | No | ||
| 115 | MPV17 | 1190133 4070427 4570577 | 16395 | -1.082 | -0.0313 | No | ||
| 116 | IMMT | 870450 4280438 | 16562 | -1.245 | -0.0356 | No | ||
| 117 | MRPL51 | 540670 1580026 | 16652 | -1.334 | -0.0353 | No | ||
| 118 | NDUFA2 | 6020100 | 16675 | -1.350 | -0.0313 | No | ||
| 119 | MRPS21 | 2650008 | 16815 | -1.529 | -0.0330 | No | ||
| 120 | NDUFA13 | 6220021 | 16843 | -1.559 | -0.0285 | No | ||
| 121 | PMPCA | 4280132 6840408 | 17004 | -1.731 | -0.0306 | No | ||
| 122 | RAF1 | 1770600 | 17308 | -2.124 | -0.0389 | No | ||
| 123 | TIMM44 | 4670746 4730050 | 17357 | -2.197 | -0.0331 | No | ||
| 124 | RHOT1 | 6760692 | 17372 | -2.221 | -0.0254 | No | ||
| 125 | ALAS2 | 6550176 | 18053 | -3.653 | -0.0483 | No | ||
| 126 | ACADM | 460112 3390113 | 18158 | -4.046 | -0.0385 | No | ||
| 127 | NDUFS4 | 510142 1240148 4760253 | 18236 | -4.416 | -0.0258 | No | ||
| 128 | MCL1 | 1660672 | 18445 | -5.818 | -0.0149 | No | ||
| 129 | CASP7 | 4210156 | 18505 | -6.324 | 0.0060 | No |

