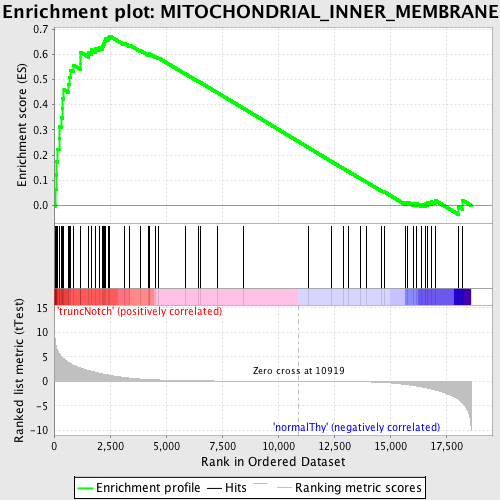
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | truncNotch |
| GeneSet | MITOCHONDRIAL_INNER_MEMBRANE |
| Enrichment Score (ES) | 0.67224324 |
| Normalized Enrichment Score (NES) | 1.6134958 |
| Nominal p-value | 0.0016778524 |
| FDR q-value | 0.14829582 |
| FWER p-Value | 0.72 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TIMM50 | 1500139 | 53 | 8.098 | 0.0640 | Yes | ||
| 2 | PHB2 | 1230050 | 91 | 7.136 | 0.1208 | Yes | ||
| 3 | TIMM13 | 2450601 | 115 | 6.699 | 0.1749 | Yes | ||
| 4 | ATP5G2 | 840068 | 168 | 6.154 | 0.2229 | Yes | ||
| 5 | BCS1L | 5360102 | 221 | 5.735 | 0.2674 | Yes | ||
| 6 | ATP5G1 | 5290019 | 253 | 5.555 | 0.3115 | Yes | ||
| 7 | ATP5E | 4120411 | 333 | 4.999 | 0.3485 | Yes | ||
| 8 | UQCRC1 | 520053 | 374 | 4.792 | 0.3859 | Yes | ||
| 9 | SLC25A1 | 4050402 | 388 | 4.747 | 0.4244 | Yes | ||
| 10 | SLC25A3 | 670112 | 439 | 4.578 | 0.4594 | Yes | ||
| 11 | ATP5D | 6550167 | 642 | 3.882 | 0.4806 | Yes | ||
| 12 | TIMM10 | 610070 1450019 | 680 | 3.769 | 0.5097 | Yes | ||
| 13 | NDUFS2 | 4850020 6200402 | 742 | 3.628 | 0.5363 | Yes | ||
| 14 | NDUFV1 | 5420369 | 859 | 3.313 | 0.5574 | Yes | ||
| 15 | SURF1 | 5130048 | 1170 | 2.751 | 0.5634 | Yes | ||
| 16 | NDUFS3 | 1690136 5340528 | 1172 | 2.750 | 0.5860 | Yes | ||
| 17 | NDUFS8 | 4150100 | 1174 | 2.747 | 0.6086 | Yes | ||
| 18 | ATP5O | 7000398 | 1544 | 2.218 | 0.6071 | Yes | ||
| 19 | SDHD | 2570128 | 1664 | 2.059 | 0.6176 | Yes | ||
| 20 | PPOX | 2640678 3850102 | 1854 | 1.867 | 0.6229 | Yes | ||
| 21 | NDUFS7 | 1740142 | 2018 | 1.698 | 0.6281 | Yes | ||
| 22 | NDUFA6 | 3990348 | 2161 | 1.527 | 0.6330 | Yes | ||
| 23 | ATP5J | 4210100 6900288 | 2216 | 1.484 | 0.6424 | Yes | ||
| 24 | TIMM17A | 130092 | 2262 | 1.446 | 0.6519 | Yes | ||
| 25 | ATP5A1 | 1990722 | 2302 | 1.410 | 0.6614 | Yes | ||
| 26 | SLC25A11 | 130097 770685 | 2424 | 1.283 | 0.6655 | Yes | ||
| 27 | COX15 | 2360402 | 2485 | 1.215 | 0.6722 | Yes | ||
| 28 | ABCB7 | 4150324 | 3130 | 0.794 | 0.6441 | No | ||
| 29 | TIMM17B | 6040400 | 3370 | 0.659 | 0.6366 | No | ||
| 30 | SLC25A15 | 4640368 5550064 | 3838 | 0.479 | 0.6154 | No | ||
| 31 | TIMM9 | 2450484 | 4219 | 0.376 | 0.5981 | No | ||
| 32 | ATP5C1 | 5910601 | 4241 | 0.371 | 0.6000 | No | ||
| 33 | TIMM23 | 60288 7040133 | 4251 | 0.367 | 0.6025 | No | ||
| 34 | NDUFA9 | 2260280 | 4528 | 0.311 | 0.5902 | No | ||
| 35 | HSD3B1 | 3060672 | 4668 | 0.287 | 0.5851 | No | ||
| 36 | HSD3B2 | 70068 770037 4120114 4230170 5690333 | 5850 | 0.148 | 0.5227 | No | ||
| 37 | UQCRB | 1450047 | 6430 | 0.112 | 0.4924 | No | ||
| 38 | ATP5G3 | 4230551 | 6523 | 0.107 | 0.4883 | No | ||
| 39 | UCP3 | 4200128 4210148 | 7303 | 0.074 | 0.4470 | No | ||
| 40 | HCCS | 670397 3170341 | 8437 | 0.044 | 0.3863 | No | ||
| 41 | PHB | 2260739 | 11377 | -0.008 | 0.2280 | No | ||
| 42 | SDHA | 7000056 | 12362 | -0.029 | 0.1752 | No | ||
| 43 | NDUFS1 | 2650253 | 12928 | -0.045 | 0.1451 | No | ||
| 44 | OPA1 | 2030433 3520390 4730537 | 13121 | -0.052 | 0.1352 | No | ||
| 45 | MRPL32 | 110041 | 13682 | -0.083 | 0.1057 | No | ||
| 46 | COX6B2 | 1340010 4120039 | 13952 | -0.104 | 0.0920 | No | ||
| 47 | NDUFAB1 | 1980091 | 14627 | -0.205 | 0.0574 | No | ||
| 48 | NNT | 540253 1170471 5550092 6760397 | 14727 | -0.231 | 0.0540 | No | ||
| 49 | SLC25A22 | 2360050 | 15689 | -0.632 | 0.0074 | No | ||
| 50 | TIMM8B | 1190364 | 15698 | -0.639 | 0.0123 | No | ||
| 51 | NDUFA1 | 6200411 | 15796 | -0.687 | 0.0127 | No | ||
| 52 | ATP5B | 3870138 | 16029 | -0.841 | 0.0071 | No | ||
| 53 | UQCRH | 4210463 | 16167 | -0.943 | 0.0075 | No | ||
| 54 | MPV17 | 1190133 4070427 4570577 | 16395 | -1.082 | 0.0042 | No | ||
| 55 | IMMT | 870450 4280438 | 16562 | -1.245 | 0.0056 | No | ||
| 56 | NDUFA2 | 6020100 | 16675 | -1.350 | 0.0107 | No | ||
| 57 | NDUFA13 | 6220021 | 16843 | -1.559 | 0.0145 | No | ||
| 58 | PMPCA | 4280132 6840408 | 17004 | -1.731 | 0.0202 | No | ||
| 59 | ALAS2 | 6550176 | 18053 | -3.653 | -0.0061 | No | ||
| 60 | NDUFS4 | 510142 1240148 4760253 | 18236 | -4.416 | 0.0205 | No |

