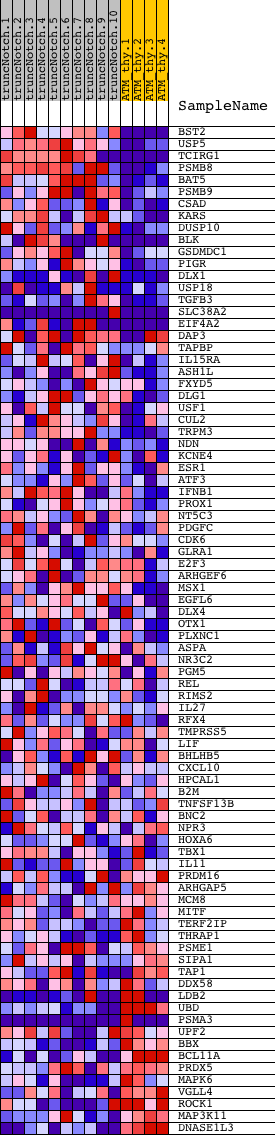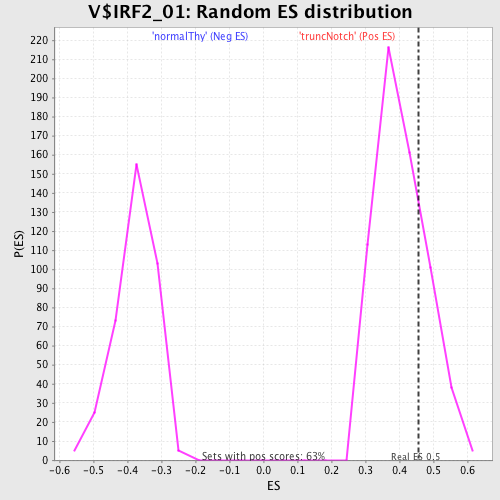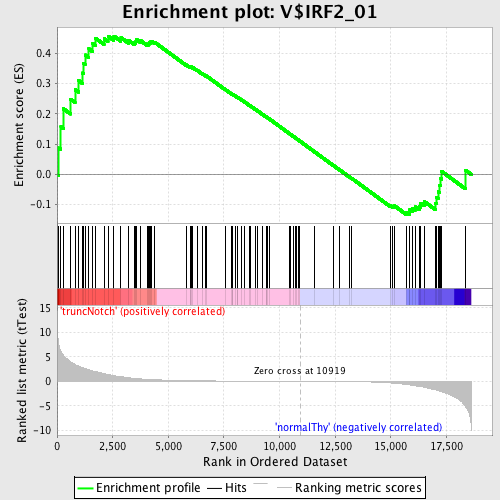
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | truncNotch |
| GeneSet | V$IRF2_01 |
| Enrichment Score (ES) | 0.45744473 |
| Normalized Enrichment Score (NES) | 1.1313415 |
| Nominal p-value | 0.23659305 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | BST2 | 6760487 | 73 | 7.401 | 0.0875 | Yes | ||
| 2 | USP5 | 4850017 | 171 | 6.090 | 0.1575 | Yes | ||
| 3 | TCIRG1 | 4590524 5910079 6900619 | 283 | 5.312 | 0.2172 | Yes | ||
| 4 | PSMB8 | 2850707 | 608 | 3.967 | 0.2487 | Yes | ||
| 5 | BAT5 | 5720687 | 809 | 3.434 | 0.2803 | Yes | ||
| 6 | PSMB9 | 6980471 | 963 | 3.106 | 0.3105 | Yes | ||
| 7 | CSAD | 770215 | 1140 | 2.798 | 0.3355 | Yes | ||
| 8 | KARS | 6520575 | 1185 | 2.730 | 0.3669 | Yes | ||
| 9 | DUSP10 | 2850673 3360064 | 1260 | 2.607 | 0.3951 | Yes | ||
| 10 | BLK | 1940128 5390053 | 1427 | 2.363 | 0.4154 | Yes | ||
| 11 | GSDMDC1 | 4010092 5050091 | 1602 | 2.134 | 0.4323 | Yes | ||
| 12 | PIGR | 6520441 | 1718 | 2.009 | 0.4509 | Yes | ||
| 13 | DLX1 | 360168 | 2111 | 1.576 | 0.4493 | Yes | ||
| 14 | USP18 | 4010110 | 2289 | 1.418 | 0.4572 | Yes | ||
| 15 | TGFB3 | 1070041 | 2553 | 1.165 | 0.4574 | Yes | ||
| 16 | SLC38A2 | 1470242 3800026 | 2869 | 0.969 | 0.4524 | No | ||
| 17 | EIF4A2 | 1170494 1740711 2850504 | 3212 | 0.749 | 0.4432 | No | ||
| 18 | DAP3 | 1660528 2120039 5420593 | 3482 | 0.611 | 0.4363 | No | ||
| 19 | TAPBP | 1980110 | 3539 | 0.586 | 0.4405 | No | ||
| 20 | IL15RA | 2570270 3190711 6100091 6100270 | 3558 | 0.580 | 0.4467 | No | ||
| 21 | ASH1L | 2510402 5360687 6650685 | 3750 | 0.509 | 0.4427 | No | ||
| 22 | FXYD5 | 4060301 6590400 | 4082 | 0.410 | 0.4299 | No | ||
| 23 | DLG1 | 2630091 6020286 | 4123 | 0.399 | 0.4326 | No | ||
| 24 | USF1 | 4280156 4610114 6370113 | 4151 | 0.393 | 0.4360 | No | ||
| 25 | CUL2 | 4200278 | 4198 | 0.381 | 0.4383 | No | ||
| 26 | TRPM3 | 2470110 6400731 6550047 | 4233 | 0.373 | 0.4411 | No | ||
| 27 | NDN | 5670075 | 4398 | 0.335 | 0.4363 | No | ||
| 28 | KCNE4 | 2470102 | 5807 | 0.152 | 0.3622 | No | ||
| 29 | ESR1 | 4060372 5860193 | 5979 | 0.137 | 0.3547 | No | ||
| 30 | ATF3 | 1940546 | 5987 | 0.137 | 0.3560 | No | ||
| 31 | IFNB1 | 1400142 | 6020 | 0.135 | 0.3560 | No | ||
| 32 | PROX1 | 4210193 | 6104 | 0.129 | 0.3531 | No | ||
| 33 | NT5C3 | 1570075 | 6297 | 0.118 | 0.3442 | No | ||
| 34 | PDGFC | 4200133 | 6545 | 0.106 | 0.3322 | No | ||
| 35 | CDK6 | 4920253 | 6677 | 0.099 | 0.3263 | No | ||
| 36 | GLRA1 | 610575 | 6707 | 0.098 | 0.3260 | No | ||
| 37 | E2F3 | 50162 460180 | 7547 | 0.066 | 0.2815 | No | ||
| 38 | ARHGEF6 | 4780164 6380450 | 7844 | 0.058 | 0.2663 | No | ||
| 39 | MSX1 | 2650309 | 7864 | 0.058 | 0.2660 | No | ||
| 40 | EGFL6 | 3940181 | 7884 | 0.057 | 0.2656 | No | ||
| 41 | DLX4 | 50215 6650059 | 8033 | 0.054 | 0.2583 | No | ||
| 42 | OTX1 | 2510215 | 8100 | 0.051 | 0.2554 | No | ||
| 43 | PLXNC1 | 5860315 | 8106 | 0.051 | 0.2558 | No | ||
| 44 | ASPA | 3290400 | 8297 | 0.047 | 0.2461 | No | ||
| 45 | NR3C2 | 2480324 6400093 | 8406 | 0.045 | 0.2408 | No | ||
| 46 | PGM5 | 2340446 | 8648 | 0.039 | 0.2283 | No | ||
| 47 | REL | 360707 | 8670 | 0.039 | 0.2276 | No | ||
| 48 | RIMS2 | 670725 | 8917 | 0.034 | 0.2148 | No | ||
| 49 | IL27 | 1990324 | 9000 | 0.032 | 0.2108 | No | ||
| 50 | RFX4 | 3710100 5340215 | 9225 | 0.028 | 0.1990 | No | ||
| 51 | TMPRSS5 | 4060068 | 9391 | 0.025 | 0.1904 | No | ||
| 52 | LIF | 1410373 2060129 3990338 | 9408 | 0.025 | 0.1899 | No | ||
| 53 | BHLHB5 | 6510520 | 9463 | 0.024 | 0.1873 | No | ||
| 54 | CXCL10 | 2450408 | 9533 | 0.023 | 0.1838 | No | ||
| 55 | HPCAL1 | 1980129 2350348 | 10427 | 0.009 | 0.1357 | No | ||
| 56 | B2M | 5080332 5130059 | 10512 | 0.007 | 0.1313 | No | ||
| 57 | TNFSF13B | 1940438 6510441 | 10641 | 0.005 | 0.1245 | No | ||
| 58 | BNC2 | 4810603 | 10700 | 0.004 | 0.1214 | No | ||
| 59 | NPR3 | 1580239 | 10768 | 0.002 | 0.1178 | No | ||
| 60 | HOXA6 | 2340333 | 10847 | 0.001 | 0.1136 | No | ||
| 61 | TBX1 | 3710133 6590121 | 10914 | 0.000 | 0.1100 | No | ||
| 62 | IL11 | 1740398 | 11588 | -0.012 | 0.0739 | No | ||
| 63 | PRDM16 | 4120541 | 12423 | -0.030 | 0.0292 | No | ||
| 64 | ARHGAP5 | 2510619 3360035 | 12688 | -0.037 | 0.0154 | No | ||
| 65 | MCM8 | 6130743 | 13120 | -0.052 | -0.0072 | No | ||
| 66 | MITF | 380056 | 13239 | -0.057 | -0.0128 | No | ||
| 67 | TERF2IP | 580010 870364 2190358 | 14970 | -0.303 | -0.1024 | No | ||
| 68 | THRAP1 | 2470121 | 15092 | -0.343 | -0.1047 | No | ||
| 69 | PSME1 | 450193 4480035 | 15166 | -0.373 | -0.1041 | No | ||
| 70 | SIPA1 | 5220687 | 15688 | -0.632 | -0.1244 | No | ||
| 71 | TAP1 | 4050047 | 15841 | -0.710 | -0.1238 | No | ||
| 72 | DDX58 | 2450075 | 15855 | -0.722 | -0.1156 | No | ||
| 73 | LDB2 | 5670441 6110170 | 15974 | -0.790 | -0.1122 | No | ||
| 74 | UBD | 5570632 | 16090 | -0.890 | -0.1074 | No | ||
| 75 | PSMA3 | 5900047 7040161 | 16279 | -1.000 | -0.1052 | No | ||
| 76 | UPF2 | 4810471 | 16342 | -1.038 | -0.0957 | No | ||
| 77 | BBX | 6520112 | 16501 | -1.192 | -0.0895 | No | ||
| 78 | BCL11A | 6860369 | 17005 | -1.733 | -0.0952 | No | ||
| 79 | PRDX5 | 1660592 2030091 | 17047 | -1.772 | -0.0755 | No | ||
| 80 | MAPK6 | 6760520 | 17131 | -1.878 | -0.0568 | No | ||
| 81 | VGLL4 | 6860463 | 17197 | -1.955 | -0.0362 | No | ||
| 82 | ROCK1 | 130044 | 17232 | -2.008 | -0.0132 | No | ||
| 83 | MAP3K11 | 7000039 | 17284 | -2.094 | 0.0099 | No | ||
| 84 | DNASE1L3 | 670086 | 18347 | -5.008 | 0.0145 | No |