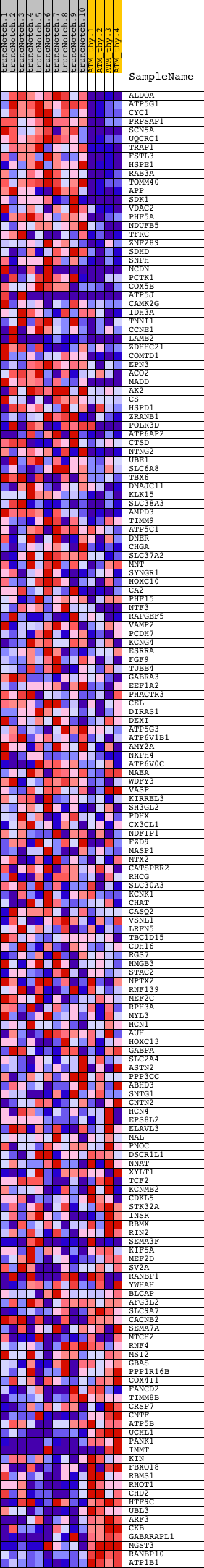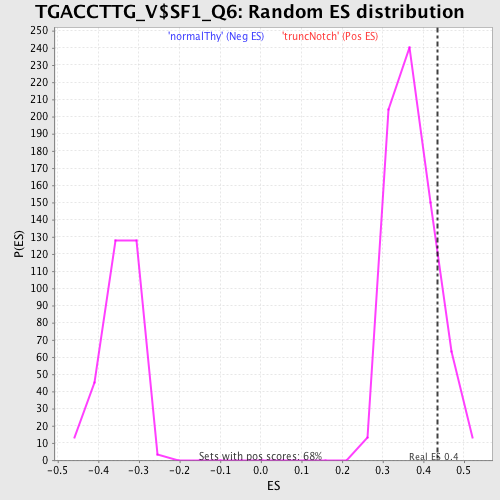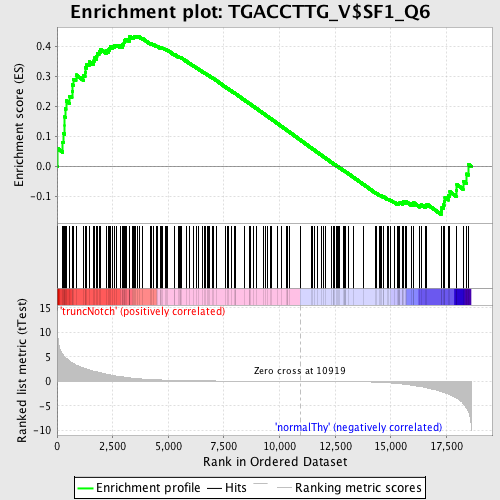
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | truncNotch |
| GeneSet | TGACCTTG_V$SF1_Q6 |
| Enrichment Score (ES) | 0.4346301 |
| Normalized Enrichment Score (NES) | 1.1687088 |
| Nominal p-value | 0.1317716 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ALDOA | 6290672 | 14 | 10.281 | 0.0603 | Yes | ||
| 2 | ATP5G1 | 5290019 | 253 | 5.555 | 0.0803 | Yes | ||
| 3 | CYC1 | 2850546 | 286 | 5.279 | 0.1099 | Yes | ||
| 4 | PRPSAP1 | 6980494 | 344 | 4.933 | 0.1361 | Yes | ||
| 5 | SCN5A | 4070017 | 346 | 4.931 | 0.1653 | Yes | ||
| 6 | UQCRC1 | 520053 | 374 | 4.792 | 0.1923 | Yes | ||
| 7 | TRAP1 | 6040168 | 400 | 4.685 | 0.2188 | Yes | ||
| 8 | FSTL3 | 5420279 | 565 | 4.151 | 0.2345 | Yes | ||
| 9 | HSPE1 | 3170438 | 670 | 3.793 | 0.2514 | Yes | ||
| 10 | RAB3A | 3940288 | 694 | 3.735 | 0.2723 | Yes | ||
| 11 | TOMM40 | 6110017 | 745 | 3.621 | 0.2911 | Yes | ||
| 12 | APP | 2510053 | 850 | 3.332 | 0.3052 | Yes | ||
| 13 | SDK1 | 2850139 | 1194 | 2.720 | 0.3028 | Yes | ||
| 14 | VDAC2 | 7000215 | 1256 | 2.610 | 0.3150 | Yes | ||
| 15 | PHF5A | 2690519 | 1268 | 2.591 | 0.3298 | Yes | ||
| 16 | NDUFB5 | 110685 | 1334 | 2.493 | 0.3410 | Yes | ||
| 17 | TFRC | 4050551 | 1439 | 2.350 | 0.3494 | Yes | ||
| 18 | ZNF289 | 3610390 3870711 3940138 5340373 | 1626 | 2.106 | 0.3518 | Yes | ||
| 19 | SDHD | 2570128 | 1664 | 2.059 | 0.3620 | Yes | ||
| 20 | SNPH | 2680484 | 1771 | 1.958 | 0.3679 | Yes | ||
| 21 | NCDN | 50040 1980739 3520603 | 1793 | 1.942 | 0.3782 | Yes | ||
| 22 | PCTK1 | 4560079 | 1885 | 1.833 | 0.3842 | Yes | ||
| 23 | COX5B | 6550670 | 1951 | 1.765 | 0.3911 | Yes | ||
| 24 | ATP5J | 4210100 6900288 | 2216 | 1.484 | 0.3856 | Yes | ||
| 25 | CAMK2G | 630097 | 2313 | 1.402 | 0.3888 | Yes | ||
| 26 | IDH3A | 460332 | 2364 | 1.357 | 0.3941 | Yes | ||
| 27 | TNNI1 | 3710040 | 2406 | 1.306 | 0.3996 | Yes | ||
| 28 | CCNE1 | 110064 | 2502 | 1.203 | 0.4016 | Yes | ||
| 29 | LAMB2 | 4920184 | 2587 | 1.141 | 0.4038 | Yes | ||
| 30 | ZDHHC21 | 2370338 | 2685 | 1.062 | 0.4049 | Yes | ||
| 31 | COMTD1 | 1170193 | 2841 | 0.986 | 0.4023 | Yes | ||
| 32 | EPN3 | 2350592 | 2933 | 0.930 | 0.4029 | Yes | ||
| 33 | ACO2 | 4230600 | 2940 | 0.926 | 0.4081 | Yes | ||
| 34 | MADD | 6100725 7000088 | 3001 | 0.881 | 0.4101 | Yes | ||
| 35 | AK2 | 4590458 4760195 6110097 | 3006 | 0.878 | 0.4151 | Yes | ||
| 36 | CS | 5080600 | 3021 | 0.868 | 0.4195 | Yes | ||
| 37 | HSPD1 | 60097 2030451 | 3056 | 0.843 | 0.4226 | Yes | ||
| 38 | ZRANB1 | 510373 4540278 | 3126 | 0.798 | 0.4236 | Yes | ||
| 39 | POLR3D | 580037 6420615 | 3240 | 0.729 | 0.4218 | Yes | ||
| 40 | ATP6AP2 | 4010059 5080315 7100347 6660484 | 3251 | 0.722 | 0.4256 | Yes | ||
| 41 | CTSD | 1070195 | 3258 | 0.717 | 0.4295 | Yes | ||
| 42 | NTNG2 | 1770438 6380408 | 3264 | 0.715 | 0.4335 | Yes | ||
| 43 | UBE1 | 4150139 7050463 | 3393 | 0.649 | 0.4304 | Yes | ||
| 44 | SLC6A8 | 3940341 | 3453 | 0.622 | 0.4309 | Yes | ||
| 45 | TBX6 | 4730139 6840647 | 3469 | 0.614 | 0.4337 | Yes | ||
| 46 | DNAJC11 | 360465 3390215 | 3518 | 0.593 | 0.4346 | Yes | ||
| 47 | KLK15 | 6350086 | 3625 | 0.546 | 0.4321 | No | ||
| 48 | SLC38A3 | 5340142 | 3682 | 0.531 | 0.4322 | No | ||
| 49 | AMPD3 | 610369 3360528 | 3853 | 0.475 | 0.4258 | No | ||
| 50 | TIMM9 | 2450484 | 4219 | 0.376 | 0.4083 | No | ||
| 51 | ATP5C1 | 5910601 | 4241 | 0.371 | 0.4094 | No | ||
| 52 | DNER | 3610170 | 4350 | 0.342 | 0.4055 | No | ||
| 53 | CHGA | 1990056 6550463 | 4457 | 0.325 | 0.4017 | No | ||
| 54 | SLC37A2 | 2970750 | 4505 | 0.315 | 0.4010 | No | ||
| 55 | MNT | 4920575 | 4645 | 0.292 | 0.3952 | No | ||
| 56 | SYNGR1 | 5130452 6650056 | 4648 | 0.291 | 0.3969 | No | ||
| 57 | HOXC10 | 5360170 | 4671 | 0.286 | 0.3974 | No | ||
| 58 | CA2 | 1660113 1660600 | 4747 | 0.273 | 0.3949 | No | ||
| 59 | PHF15 | 6550066 6860673 | 4868 | 0.256 | 0.3899 | No | ||
| 60 | NTF3 | 5890131 | 4896 | 0.251 | 0.3900 | No | ||
| 61 | RAPGEF5 | 1500603 3450427 6130056 | 4962 | 0.244 | 0.3879 | No | ||
| 62 | VAMP2 | 2100100 | 5268 | 0.207 | 0.3726 | No | ||
| 63 | PCDH7 | 450398 4150040 | 5284 | 0.205 | 0.3730 | No | ||
| 64 | KCNG4 | 1850736 5050113 | 5441 | 0.187 | 0.3656 | No | ||
| 65 | ESRRA | 6550242 | 5482 | 0.182 | 0.3646 | No | ||
| 66 | FGF9 | 1050195 | 5531 | 0.178 | 0.3630 | No | ||
| 67 | TUBB4 | 5310681 | 5551 | 0.176 | 0.3630 | No | ||
| 68 | GABRA3 | 4810142 | 5573 | 0.175 | 0.3629 | No | ||
| 69 | EEF1A2 | 2260162 5290086 | 5795 | 0.153 | 0.3519 | No | ||
| 70 | PHACTR3 | 3850435 5900445 | 5968 | 0.138 | 0.3433 | No | ||
| 71 | CEL | 2640278 | 6114 | 0.129 | 0.3363 | No | ||
| 72 | DIRAS1 | 2640270 | 6266 | 0.120 | 0.3288 | No | ||
| 73 | DEXI | 6220358 6550685 | 6355 | 0.115 | 0.3247 | No | ||
| 74 | ATP5G3 | 4230551 | 6523 | 0.107 | 0.3163 | No | ||
| 75 | ATP6V1B1 | 6100181 | 6627 | 0.102 | 0.3113 | No | ||
| 76 | AMY2A | 580138 | 6674 | 0.099 | 0.3094 | No | ||
| 77 | NXPH4 | 3830500 | 6759 | 0.096 | 0.3054 | No | ||
| 78 | ATP6V0C | 1780609 | 6801 | 0.094 | 0.3037 | No | ||
| 79 | MAEA | 1170392 | 6866 | 0.092 | 0.3008 | No | ||
| 80 | WDFY3 | 3610041 4560333 4570273 | 6987 | 0.086 | 0.2948 | No | ||
| 81 | VASP | 7050500 | 7044 | 0.083 | 0.2923 | No | ||
| 82 | KIRREL3 | 2630195 | 7148 | 0.080 | 0.2872 | No | ||
| 83 | SH3GL2 | 1990176 3830494 | 7587 | 0.065 | 0.2638 | No | ||
| 84 | PDHX | 870315 | 7638 | 0.064 | 0.2615 | No | ||
| 85 | CX3CL1 | 3990707 | 7685 | 0.063 | 0.2594 | No | ||
| 86 | NDFIP1 | 1980402 | 7829 | 0.059 | 0.2520 | No | ||
| 87 | FZD9 | 5360136 | 7959 | 0.055 | 0.2453 | No | ||
| 88 | MASP1 | 1780619 2900066 | 8003 | 0.054 | 0.2433 | No | ||
| 89 | MTX2 | 6660288 | 8025 | 0.054 | 0.2425 | No | ||
| 90 | CATSPER2 | 2940403 | 8424 | 0.044 | 0.2212 | No | ||
| 91 | RHCG | 6550309 | 8626 | 0.040 | 0.2105 | No | ||
| 92 | SLC30A3 | 1850451 | 8644 | 0.039 | 0.2098 | No | ||
| 93 | KCNK1 | 1050068 | 8676 | 0.039 | 0.2084 | No | ||
| 94 | CHAT | 6840603 | 8811 | 0.036 | 0.2013 | No | ||
| 95 | CASQ2 | 3170136 | 8982 | 0.032 | 0.1923 | No | ||
| 96 | VSNL1 | 3120017 | 9270 | 0.028 | 0.1769 | No | ||
| 97 | LRFN5 | 6650088 | 9354 | 0.026 | 0.1725 | No | ||
| 98 | TBC1D15 | 870673 | 9458 | 0.024 | 0.1671 | No | ||
| 99 | CDH16 | 1450129 | 9586 | 0.022 | 0.1604 | No | ||
| 100 | RGS7 | 3390044 | 9638 | 0.021 | 0.1577 | No | ||
| 101 | HMGB3 | 2940168 | 9924 | 0.016 | 0.1424 | No | ||
| 102 | STAC2 | 7100022 | 10105 | 0.014 | 0.1327 | No | ||
| 103 | NPTX2 | 70021 | 10296 | 0.011 | 0.1225 | No | ||
| 104 | RNF139 | 2650114 3610450 | 10360 | 0.010 | 0.1191 | No | ||
| 105 | MEF2C | 670025 780338 | 10374 | 0.009 | 0.1184 | No | ||
| 106 | RPH3A | 2190156 | 10434 | 0.009 | 0.1153 | No | ||
| 107 | MYL3 | 6040563 | 10945 | -0.000 | 0.0877 | No | ||
| 108 | HCN1 | 5130458 | 11429 | -0.009 | 0.0615 | No | ||
| 109 | AUH | 5570152 | 11459 | -0.009 | 0.0600 | No | ||
| 110 | HOXC13 | 4070722 | 11553 | -0.011 | 0.0550 | No | ||
| 111 | GABPA | 2360017 4780301 4850279 | 11719 | -0.015 | 0.0462 | No | ||
| 112 | SLC2A4 | 540441 | 11889 | -0.018 | 0.0371 | No | ||
| 113 | ASTN2 | 2570136 6940040 | 11979 | -0.020 | 0.0324 | No | ||
| 114 | PPP3CC | 2450139 | 12057 | -0.021 | 0.0284 | No | ||
| 115 | ABHD3 | 2370091 | 12324 | -0.027 | 0.0141 | No | ||
| 116 | SNTG1 | 6770373 | 12436 | -0.030 | 0.0083 | No | ||
| 117 | CNTN2 | 2760204 | 12442 | -0.031 | 0.0082 | No | ||
| 118 | HCN4 | 1050026 | 12461 | -0.031 | 0.0074 | No | ||
| 119 | EPS8L2 | 2850528 | 12563 | -0.034 | 0.0021 | No | ||
| 120 | ELAVL3 | 2850014 | 12594 | -0.035 | 0.0007 | No | ||
| 121 | MAL | 4280487 4590239 | 12634 | -0.035 | -0.0012 | No | ||
| 122 | PNOC | 1990114 | 12703 | -0.038 | -0.0047 | No | ||
| 123 | DSCR1L1 | 940438 | 12856 | -0.042 | -0.0127 | No | ||
| 124 | NNAT | 4920053 5130253 | 12904 | -0.044 | -0.0150 | No | ||
| 125 | XYLT1 | 6550403 | 12927 | -0.045 | -0.0159 | No | ||
| 126 | TCF2 | 870338 5050632 | 12959 | -0.046 | -0.0173 | No | ||
| 127 | KCNMB2 | 3780128 4760136 | 12973 | -0.046 | -0.0177 | No | ||
| 128 | CDKL5 | 2450070 | 13087 | -0.051 | -0.0235 | No | ||
| 129 | STK32A | 5890332 | 13339 | -0.062 | -0.0368 | No | ||
| 130 | INSR | 1190504 | 13765 | -0.089 | -0.0593 | No | ||
| 131 | RBMX | 7100162 2900541 | 14308 | -0.145 | -0.0878 | No | ||
| 132 | RIN2 | 2450184 | 14355 | -0.154 | -0.0894 | No | ||
| 133 | SEMA3F | 5420601 6770731 | 14497 | -0.178 | -0.0960 | No | ||
| 134 | KIF5A | 510435 | 14541 | -0.187 | -0.0972 | No | ||
| 135 | MEF2D | 5690576 | 14566 | -0.193 | -0.0974 | No | ||
| 136 | SV2A | 1450039 5570685 | 14674 | -0.216 | -0.1019 | No | ||
| 137 | RANBP1 | 430215 1090180 | 14677 | -0.217 | -0.1007 | No | ||
| 138 | YWHAH | 1660133 2810053 | 14856 | -0.272 | -0.1087 | No | ||
| 139 | BLCAP | 3990253 | 14892 | -0.282 | -0.1090 | No | ||
| 140 | AFG3L2 | 5130332 | 14976 | -0.306 | -0.1116 | No | ||
| 141 | SLC9A7 | 5360301 | 15156 | -0.365 | -0.1192 | No | ||
| 142 | CACNB2 | 1500095 7330707 | 15282 | -0.415 | -0.1235 | No | ||
| 143 | SEMA7A | 2630239 5050593 | 15335 | -0.437 | -0.1237 | No | ||
| 144 | MTCH2 | 3780603 6620215 | 15359 | -0.451 | -0.1223 | No | ||
| 145 | RNF4 | 580286 1400152 | 15404 | -0.479 | -0.1218 | No | ||
| 146 | MSI2 | 1190066 5270039 | 15406 | -0.480 | -0.1190 | No | ||
| 147 | GBAS | 6760278 | 15539 | -0.545 | -0.1230 | No | ||
| 148 | PPP1R16B | 3520300 | 15546 | -0.548 | -0.1200 | No | ||
| 149 | COX4I1 | 5340427 | 15548 | -0.548 | -0.1168 | No | ||
| 150 | FANCD2 | 1850280 6840348 | 15666 | -0.616 | -0.1195 | No | ||
| 151 | TIMM8B | 1190364 | 15698 | -0.639 | -0.1174 | No | ||
| 152 | CRSP7 | 4230408 | 15912 | -0.757 | -0.1245 | No | ||
| 153 | CNTF | 450128 870253 1780068 6520195 | 16019 | -0.829 | -0.1253 | No | ||
| 154 | ATP5B | 3870138 | 16029 | -0.841 | -0.1208 | No | ||
| 155 | UCHL1 | 1230066 | 16297 | -1.003 | -0.1293 | No | ||
| 156 | PANK1 | 670091 6220605 | 16372 | -1.066 | -0.1270 | No | ||
| 157 | IMMT | 870450 4280438 | 16562 | -1.245 | -0.1298 | No | ||
| 158 | KIN | 6550014 | 16619 | -1.305 | -0.1251 | No | ||
| 159 | FBXO18 | 1240731 6370541 | 17262 | -2.055 | -0.1477 | No | ||
| 160 | RBMS1 | 6400014 | 17294 | -2.106 | -0.1369 | No | ||
| 161 | RHOT1 | 6760692 | 17372 | -2.221 | -0.1279 | No | ||
| 162 | CHD2 | 4070039 | 17394 | -2.267 | -0.1156 | No | ||
| 163 | HTF9C | 3190048 4120132 | 17432 | -2.321 | -0.1038 | No | ||
| 164 | UBL3 | 6450458 | 17580 | -2.576 | -0.0965 | No | ||
| 165 | ARF3 | 3710446 | 17654 | -2.743 | -0.0842 | No | ||
| 166 | CKB | 4560632 | 17956 | -3.417 | -0.0802 | No | ||
| 167 | GABARAPL1 | 2810458 | 17968 | -3.447 | -0.0603 | No | ||
| 168 | MGST3 | 3450338 5290736 | 18250 | -4.492 | -0.0489 | No | ||
| 169 | RANBP10 | 2340184 | 18412 | -5.513 | -0.0249 | No | ||
| 170 | ATP1B1 | 3130594 | 18480 | -6.052 | 0.0074 | No |