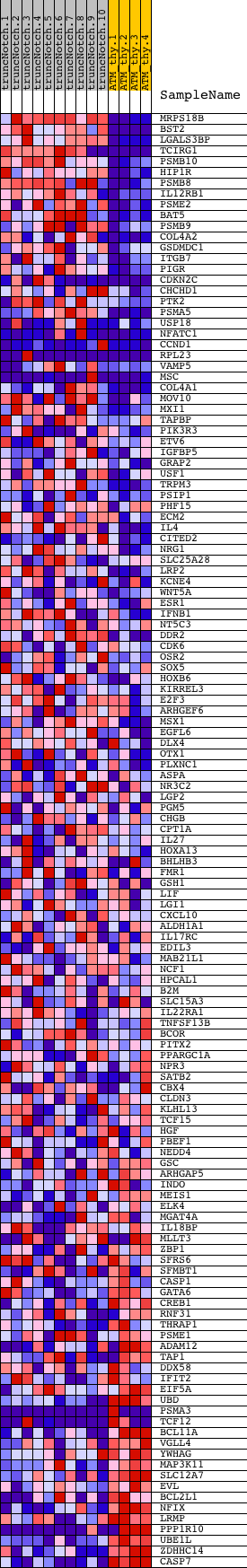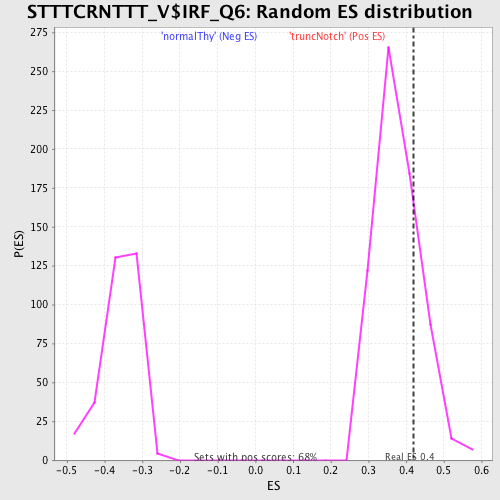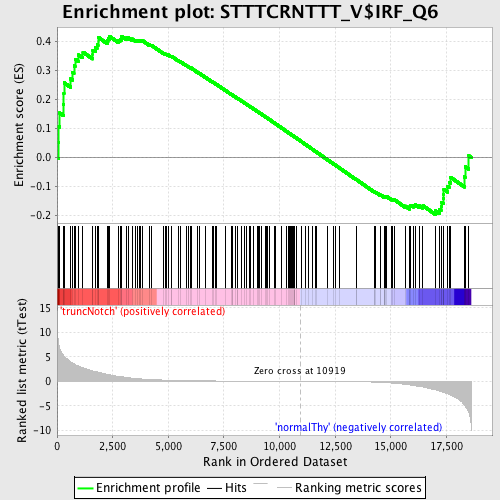
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | truncNotch |
| GeneSet | STTTCRNTTT_V$IRF_Q6 |
| Enrichment Score (ES) | 0.41865692 |
| Normalized Enrichment Score (NES) | 1.1052262 |
| Nominal p-value | 0.24300441 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MRPS18B | 5570086 | 62 | 7.759 | 0.0535 | Yes | ||
| 2 | BST2 | 6760487 | 73 | 7.401 | 0.1072 | Yes | ||
| 3 | LGALS3BP | 840692 | 117 | 6.695 | 0.1540 | Yes | ||
| 4 | TCIRG1 | 4590524 5910079 6900619 | 283 | 5.312 | 0.1840 | Yes | ||
| 5 | PSMB10 | 630504 6510039 | 303 | 5.190 | 0.2210 | Yes | ||
| 6 | HIP1R | 3120368 | 312 | 5.135 | 0.2582 | Yes | ||
| 7 | PSMB8 | 2850707 | 608 | 3.967 | 0.2713 | Yes | ||
| 8 | IL12RB1 | 2230338 5420053 | 689 | 3.748 | 0.2944 | Yes | ||
| 9 | PSME2 | 4850086 | 761 | 3.578 | 0.3168 | Yes | ||
| 10 | BAT5 | 5720687 | 809 | 3.434 | 0.3394 | Yes | ||
| 11 | PSMB9 | 6980471 | 963 | 3.106 | 0.3539 | Yes | ||
| 12 | COL4A2 | 2350619 | 1161 | 2.766 | 0.3635 | Yes | ||
| 13 | GSDMDC1 | 4010092 5050091 | 1602 | 2.134 | 0.3554 | Yes | ||
| 14 | ITGB7 | 4670619 | 1606 | 2.123 | 0.3708 | Yes | ||
| 15 | PIGR | 6520441 | 1718 | 2.009 | 0.3795 | Yes | ||
| 16 | CDKN2C | 5050750 5130148 | 1796 | 1.940 | 0.3895 | Yes | ||
| 17 | CHCHD1 | 110706 | 1841 | 1.888 | 0.4010 | Yes | ||
| 18 | PTK2 | 1780148 | 1842 | 1.887 | 0.4148 | Yes | ||
| 19 | PSMA5 | 5390537 | 2263 | 1.443 | 0.4027 | Yes | ||
| 20 | USP18 | 4010110 | 2289 | 1.418 | 0.4117 | Yes | ||
| 21 | NFATC1 | 510400 2320348 4050600 6180161 6290136 6620086 | 2365 | 1.357 | 0.4176 | Yes | ||
| 22 | CCND1 | 460524 770309 3120576 6980398 | 2748 | 1.025 | 0.4044 | Yes | ||
| 23 | RPL23 | 2260452 4850435 6450673 | 2828 | 0.989 | 0.4074 | Yes | ||
| 24 | VAMP5 | 3830300 | 2881 | 0.959 | 0.4116 | Yes | ||
| 25 | MSC | 5080139 | 2882 | 0.959 | 0.4187 | Yes | ||
| 26 | COL4A1 | 1740575 | 3106 | 0.814 | 0.4126 | No | ||
| 27 | MOV10 | 50673 430577 3870280 | 3198 | 0.757 | 0.4132 | No | ||
| 28 | MXI1 | 5050064 5130484 | 3368 | 0.660 | 0.4089 | No | ||
| 29 | TAPBP | 1980110 | 3539 | 0.586 | 0.4040 | No | ||
| 30 | PIK3R3 | 1770333 | 3593 | 0.560 | 0.4052 | No | ||
| 31 | ETV6 | 610524 | 3688 | 0.528 | 0.4040 | No | ||
| 32 | IGFBP5 | 2360592 | 3749 | 0.510 | 0.4045 | No | ||
| 33 | GRAP2 | 7100441 1410647 | 3859 | 0.474 | 0.4021 | No | ||
| 34 | USF1 | 4280156 4610114 6370113 | 4151 | 0.393 | 0.3892 | No | ||
| 35 | TRPM3 | 2470110 6400731 6550047 | 4233 | 0.373 | 0.3876 | No | ||
| 36 | PSIP1 | 1780082 3190435 5050594 | 4772 | 0.270 | 0.3604 | No | ||
| 37 | PHF15 | 6550066 6860673 | 4868 | 0.256 | 0.3572 | No | ||
| 38 | ECM2 | 1470561 | 4913 | 0.249 | 0.3566 | No | ||
| 39 | IL4 | 6020537 | 4994 | 0.240 | 0.3540 | No | ||
| 40 | CITED2 | 5670114 5130088 | 5122 | 0.223 | 0.3488 | No | ||
| 41 | NRG1 | 1050332 | 5146 | 0.221 | 0.3492 | No | ||
| 42 | SLC25A28 | 6590600 | 5436 | 0.188 | 0.3349 | No | ||
| 43 | LRP2 | 1400204 3290706 | 5562 | 0.175 | 0.3294 | No | ||
| 44 | KCNE4 | 2470102 | 5807 | 0.152 | 0.3173 | No | ||
| 45 | WNT5A | 840685 3120152 | 5891 | 0.145 | 0.3139 | No | ||
| 46 | ESR1 | 4060372 5860193 | 5979 | 0.137 | 0.3102 | No | ||
| 47 | IFNB1 | 1400142 | 6020 | 0.135 | 0.3090 | No | ||
| 48 | NT5C3 | 1570075 | 6297 | 0.118 | 0.2950 | No | ||
| 49 | DDR2 | 7050273 | 6407 | 0.112 | 0.2899 | No | ||
| 50 | CDK6 | 4920253 | 6677 | 0.099 | 0.2761 | No | ||
| 51 | OSR2 | 70711 | 6967 | 0.087 | 0.2611 | No | ||
| 52 | SOX5 | 2370576 2900167 3190128 5050528 | 7019 | 0.085 | 0.2589 | No | ||
| 53 | HOXB6 | 4210739 | 7117 | 0.081 | 0.2543 | No | ||
| 54 | KIRREL3 | 2630195 | 7148 | 0.080 | 0.2532 | No | ||
| 55 | E2F3 | 50162 460180 | 7547 | 0.066 | 0.2322 | No | ||
| 56 | ARHGEF6 | 4780164 6380450 | 7844 | 0.058 | 0.2166 | No | ||
| 57 | MSX1 | 2650309 | 7864 | 0.058 | 0.2160 | No | ||
| 58 | EGFL6 | 3940181 | 7884 | 0.057 | 0.2154 | No | ||
| 59 | DLX4 | 50215 6650059 | 8033 | 0.054 | 0.2078 | No | ||
| 60 | OTX1 | 2510215 | 8100 | 0.051 | 0.2046 | No | ||
| 61 | PLXNC1 | 5860315 | 8106 | 0.051 | 0.2047 | No | ||
| 62 | ASPA | 3290400 | 8297 | 0.047 | 0.1947 | No | ||
| 63 | NR3C2 | 2480324 6400093 | 8406 | 0.045 | 0.1892 | No | ||
| 64 | LGP2 | 4480338 | 8499 | 0.042 | 0.1846 | No | ||
| 65 | PGM5 | 2340446 | 8648 | 0.039 | 0.1768 | No | ||
| 66 | CHGB | 1980053 | 8685 | 0.039 | 0.1752 | No | ||
| 67 | CPT1A | 6350093 | 8846 | 0.035 | 0.1668 | No | ||
| 68 | IL27 | 1990324 | 9000 | 0.032 | 0.1587 | No | ||
| 69 | HOXA13 | 3120647 | 9043 | 0.032 | 0.1567 | No | ||
| 70 | BHLHB3 | 3450438 | 9084 | 0.031 | 0.1548 | No | ||
| 71 | FMR1 | 5050075 | 9171 | 0.029 | 0.1503 | No | ||
| 72 | GSH1 | 2760242 | 9383 | 0.026 | 0.1391 | No | ||
| 73 | LIF | 1410373 2060129 3990338 | 9408 | 0.025 | 0.1380 | No | ||
| 74 | LGI1 | 1850020 | 9477 | 0.024 | 0.1345 | No | ||
| 75 | CXCL10 | 2450408 | 9533 | 0.023 | 0.1317 | No | ||
| 76 | ALDH1A1 | 6520706 | 9775 | 0.019 | 0.1188 | No | ||
| 77 | IL17RC | 630164 2810593 | 9821 | 0.018 | 0.1165 | No | ||
| 78 | EDIL3 | 1240039 | 10106 | 0.014 | 0.1012 | No | ||
| 79 | MAB21L1 | 1450634 1580092 | 10315 | 0.010 | 0.0900 | No | ||
| 80 | NCF1 | 3850091 | 10422 | 0.009 | 0.0843 | No | ||
| 81 | HPCAL1 | 1980129 2350348 | 10427 | 0.009 | 0.0842 | No | ||
| 82 | B2M | 5080332 5130059 | 10512 | 0.007 | 0.0797 | No | ||
| 83 | SLC15A3 | 6650333 | 10534 | 0.007 | 0.0786 | No | ||
| 84 | IL22RA1 | 5720603 | 10559 | 0.006 | 0.0774 | No | ||
| 85 | TNFSF13B | 1940438 6510441 | 10641 | 0.005 | 0.0730 | No | ||
| 86 | BCOR | 1660452 3940053 | 10650 | 0.005 | 0.0726 | No | ||
| 87 | PITX2 | 870537 2690139 | 10665 | 0.004 | 0.0719 | No | ||
| 88 | PPARGC1A | 4670040 | 10672 | 0.004 | 0.0716 | No | ||
| 89 | NPR3 | 1580239 | 10768 | 0.002 | 0.0665 | No | ||
| 90 | SATB2 | 2470181 | 10987 | -0.001 | 0.0547 | No | ||
| 91 | CBX4 | 70332 | 11176 | -0.004 | 0.0445 | No | ||
| 92 | CLDN3 | 2570524 | 11309 | -0.007 | 0.0374 | No | ||
| 93 | KLHL13 | 6590731 | 11472 | -0.010 | 0.0287 | No | ||
| 94 | TCF15 | 1690400 | 11591 | -0.012 | 0.0224 | No | ||
| 95 | HGF | 3360593 | 11644 | -0.013 | 0.0197 | No | ||
| 96 | PBEF1 | 1090278 | 12172 | -0.024 | -0.0086 | No | ||
| 97 | NEDD4 | 460736 1190280 | 12410 | -0.030 | -0.0212 | No | ||
| 98 | GSC | 1580685 | 12496 | -0.032 | -0.0256 | No | ||
| 99 | ARHGAP5 | 2510619 3360035 | 12688 | -0.037 | -0.0356 | No | ||
| 100 | INDO | 2680390 | 13438 | -0.067 | -0.0757 | No | ||
| 101 | MEIS1 | 1400575 | 13439 | -0.067 | -0.0752 | No | ||
| 102 | ELK4 | 460494 940369 4480136 | 14249 | -0.136 | -0.1180 | No | ||
| 103 | MGAT4A | 5690471 | 14254 | -0.136 | -0.1172 | No | ||
| 104 | IL18BP | 3450128 | 14312 | -0.146 | -0.1192 | No | ||
| 105 | MLLT3 | 510204 3850722 6200088 | 14536 | -0.187 | -0.1299 | No | ||
| 106 | ZBP1 | 4210138 | 14542 | -0.188 | -0.1288 | No | ||
| 107 | SFRS6 | 60224 | 14710 | -0.226 | -0.1362 | No | ||
| 108 | SFMBT1 | 2360450 2370070 | 14729 | -0.232 | -0.1354 | No | ||
| 109 | CASP1 | 2360546 | 14735 | -0.234 | -0.1340 | No | ||
| 110 | GATA6 | 1770010 | 14763 | -0.243 | -0.1337 | No | ||
| 111 | CREB1 | 1500717 2230358 3610600 6550601 | 14820 | -0.260 | -0.1348 | No | ||
| 112 | RNF31 | 110035 | 15038 | -0.322 | -0.1442 | No | ||
| 113 | THRAP1 | 2470121 | 15092 | -0.343 | -0.1445 | No | ||
| 114 | PSME1 | 450193 4480035 | 15166 | -0.373 | -0.1457 | No | ||
| 115 | ADAM12 | 3390132 4070347 | 15641 | -0.599 | -0.1670 | No | ||
| 116 | TAP1 | 4050047 | 15841 | -0.710 | -0.1726 | No | ||
| 117 | DDX58 | 2450075 | 15855 | -0.722 | -0.1680 | No | ||
| 118 | IFIT2 | 4810735 | 15890 | -0.742 | -0.1644 | No | ||
| 119 | EIF5A | 1500059 5290022 6370026 | 16008 | -0.822 | -0.1647 | No | ||
| 120 | UBD | 5570632 | 16090 | -0.890 | -0.1625 | No | ||
| 121 | PSMA3 | 5900047 7040161 | 16279 | -1.000 | -0.1654 | No | ||
| 122 | TCF12 | 3610324 7000156 | 16445 | -1.131 | -0.1660 | No | ||
| 123 | BCL11A | 6860369 | 17005 | -1.733 | -0.1836 | No | ||
| 124 | VGLL4 | 6860463 | 17197 | -1.955 | -0.1796 | No | ||
| 125 | YWHAG | 3780341 | 17260 | -2.052 | -0.1679 | No | ||
| 126 | MAP3K11 | 7000039 | 17284 | -2.094 | -0.1538 | No | ||
| 127 | SLC12A7 | 6860079 | 17361 | -2.201 | -0.1418 | No | ||
| 128 | EVL | 1740113 | 17376 | -2.229 | -0.1262 | No | ||
| 129 | BCL2L1 | 1580452 4200152 5420484 | 17384 | -2.251 | -0.1101 | No | ||
| 130 | NFIX | 2450152 | 17552 | -2.521 | -0.1006 | No | ||
| 131 | LRMP | 6290193 | 17622 | -2.650 | -0.0849 | No | ||
| 132 | PPP1R10 | 110136 3170725 3610484 | 17683 | -2.782 | -0.0678 | No | ||
| 133 | UBE1L | 6660242 | 18312 | -4.835 | -0.0663 | No | ||
| 134 | ZDHHC14 | 3780458 | 18341 | -4.959 | -0.0315 | No | ||
| 135 | CASP7 | 4210156 | 18505 | -6.324 | 0.0060 | No |