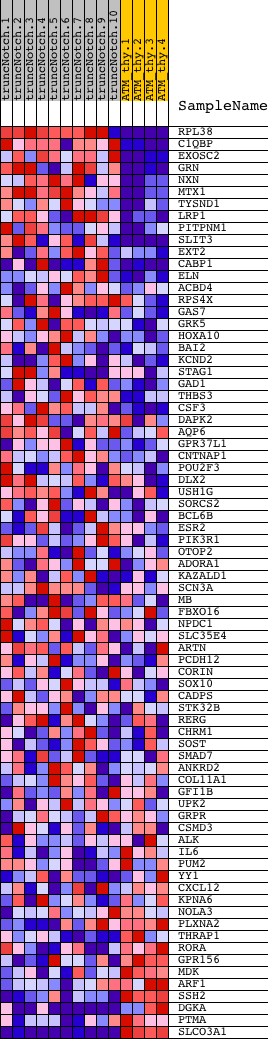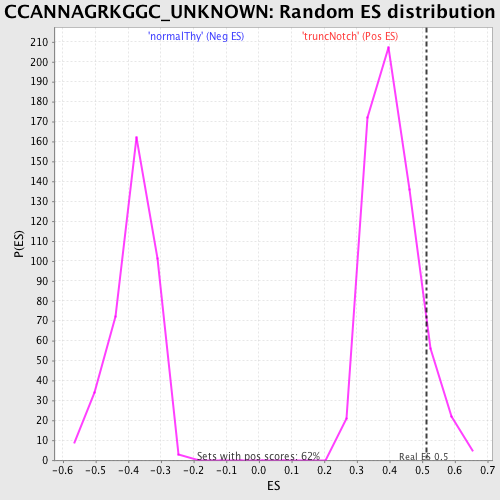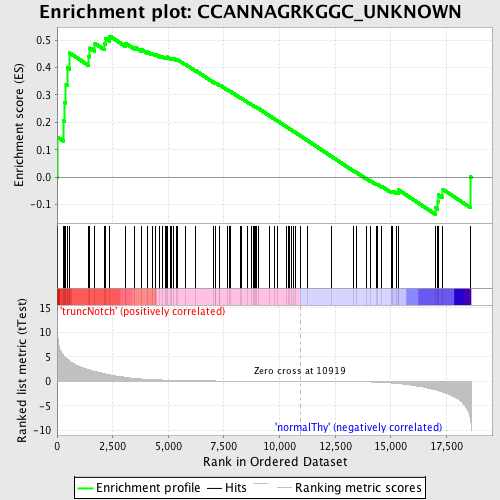
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | truncNotch |
| GeneSet | CCANNAGRKGGC_UNKNOWN |
| Enrichment Score (ES) | 0.514604 |
| Normalized Enrichment Score (NES) | 1.2549638 |
| Nominal p-value | 0.09854604 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RPL38 | 70040 4060138 6200601 | 13 | 10.322 | 0.1464 | Yes | ||
| 2 | C1QBP | 3170452 | 295 | 5.228 | 0.2057 | Yes | ||
| 3 | EXOSC2 | 6900471 | 345 | 4.933 | 0.2734 | Yes | ||
| 4 | GRN | 4050239 | 398 | 4.687 | 0.3374 | Yes | ||
| 5 | NXN | 2350180 | 450 | 4.544 | 0.3994 | Yes | ||
| 6 | MTX1 | 5910059 6550181 | 559 | 4.190 | 0.4533 | Yes | ||
| 7 | TYSND1 | 2940273 | 1403 | 2.392 | 0.4419 | Yes | ||
| 8 | LRP1 | 6270386 | 1470 | 2.314 | 0.4713 | Yes | ||
| 9 | PITPNM1 | 4540528 | 1700 | 2.028 | 0.4879 | Yes | ||
| 10 | SLIT3 | 7100132 | 2129 | 1.560 | 0.4870 | Yes | ||
| 11 | EXT2 | 6520239 | 2190 | 1.500 | 0.5051 | Yes | ||
| 12 | CABP1 | 6350750 | 2372 | 1.349 | 0.5146 | Yes | ||
| 13 | ELN | 5080347 | 3075 | 0.832 | 0.4886 | No | ||
| 14 | ACBD4 | 6980079 | 3490 | 0.607 | 0.4749 | No | ||
| 15 | RPS4X | 2690020 | 3784 | 0.500 | 0.4662 | No | ||
| 16 | GAS7 | 2120358 6450494 | 4059 | 0.417 | 0.4574 | No | ||
| 17 | GRK5 | 1940348 4670053 | 4273 | 0.360 | 0.4510 | No | ||
| 18 | HOXA10 | 6110397 7100458 | 4431 | 0.329 | 0.4473 | No | ||
| 19 | BAI2 | 380632 630113 6650170 | 4603 | 0.299 | 0.4423 | No | ||
| 20 | KCND2 | 1170128 2100112 | 4742 | 0.274 | 0.4388 | No | ||
| 21 | STAG1 | 1190300 1400722 4590100 | 4856 | 0.257 | 0.4363 | No | ||
| 22 | GAD1 | 2360035 3140167 | 4919 | 0.249 | 0.4365 | No | ||
| 23 | THBS3 | 2360465 4050176 | 4944 | 0.246 | 0.4387 | No | ||
| 24 | CSF3 | 2230193 6660707 | 5107 | 0.224 | 0.4332 | No | ||
| 25 | DAPK2 | 5890066 | 5147 | 0.220 | 0.4342 | No | ||
| 26 | AQP6 | 510736 1570484 | 5245 | 0.209 | 0.4320 | No | ||
| 27 | GPR37L1 | 5900373 | 5373 | 0.195 | 0.4279 | No | ||
| 28 | CNTNAP1 | 6650132 | 5399 | 0.193 | 0.4293 | No | ||
| 29 | POU2F3 | 2030601 | 5751 | 0.157 | 0.4126 | No | ||
| 30 | DLX2 | 2320438 4200673 | 6237 | 0.122 | 0.3882 | No | ||
| 31 | USH1G | 2940446 | 7013 | 0.085 | 0.3476 | No | ||
| 32 | SORCS2 | 6350520 | 7112 | 0.081 | 0.3435 | No | ||
| 33 | BCL6B | 60047 | 7136 | 0.080 | 0.3434 | No | ||
| 34 | ESR2 | 6220138 | 7306 | 0.074 | 0.3353 | No | ||
| 35 | PIK3R1 | 4730671 | 7309 | 0.074 | 0.3363 | No | ||
| 36 | OTOP2 | 610068 | 7677 | 0.063 | 0.3174 | No | ||
| 37 | ADORA1 | 6370520 | 7745 | 0.061 | 0.3146 | No | ||
| 38 | KAZALD1 | 380603 | 7776 | 0.060 | 0.3138 | No | ||
| 39 | SCN3A | 4070309 | 8259 | 0.048 | 0.2885 | No | ||
| 40 | MB | 730019 | 8279 | 0.047 | 0.2882 | No | ||
| 41 | FBXO16 | 6770576 | 8560 | 0.041 | 0.2737 | No | ||
| 42 | NPDC1 | 2100445 | 8718 | 0.038 | 0.2657 | No | ||
| 43 | SLC35E4 | 2370711 | 8817 | 0.036 | 0.2610 | No | ||
| 44 | ARTN | 360670 1170333 1500520 | 8851 | 0.035 | 0.2597 | No | ||
| 45 | PCDH12 | 5340044 | 8937 | 0.033 | 0.2556 | No | ||
| 46 | CORIN | 1980253 | 8946 | 0.033 | 0.2556 | No | ||
| 47 | SOX10 | 6200538 | 9059 | 0.031 | 0.2500 | No | ||
| 48 | CADPS | 2320181 | 9554 | 0.023 | 0.2237 | No | ||
| 49 | STK32B | 7000204 | 9789 | 0.019 | 0.2114 | No | ||
| 50 | RERG | 5910324 | 9892 | 0.017 | 0.2061 | No | ||
| 51 | CHRM1 | 4280619 | 10310 | 0.010 | 0.1837 | No | ||
| 52 | SOST | 1170195 | 10392 | 0.009 | 0.1795 | No | ||
| 53 | SMAD7 | 430377 | 10452 | 0.008 | 0.1764 | No | ||
| 54 | ANKRD2 | 6590427 | 10513 | 0.007 | 0.1733 | No | ||
| 55 | COL11A1 | 2100300 2810136 4200706 4810524 | 10629 | 0.005 | 0.1672 | No | ||
| 56 | GFI1B | 4570131 | 10704 | 0.004 | 0.1632 | No | ||
| 57 | UPK2 | 60176 | 10929 | -0.000 | 0.1512 | No | ||
| 58 | GRPR | 6020170 | 11241 | -0.005 | 0.1345 | No | ||
| 59 | CSMD3 | 1660427 1940687 | 12334 | -0.027 | 0.0760 | No | ||
| 60 | ALK | 730008 | 13299 | -0.060 | 0.0248 | No | ||
| 61 | IL6 | 380133 | 13465 | -0.069 | 0.0169 | No | ||
| 62 | PUM2 | 4200441 5910446 | 13889 | -0.097 | -0.0045 | No | ||
| 63 | YY1 | 6130451 | 14080 | -0.115 | -0.0131 | No | ||
| 64 | CXCL12 | 580546 4150750 4570068 | 14346 | -0.152 | -0.0253 | No | ||
| 65 | KPNA6 | 1500064 | 14387 | -0.158 | -0.0252 | No | ||
| 66 | NOLA3 | 6940687 | 14589 | -0.197 | -0.0332 | No | ||
| 67 | PLXNA2 | 6450132 | 15025 | -0.319 | -0.0521 | No | ||
| 68 | THRAP1 | 2470121 | 15092 | -0.343 | -0.0508 | No | ||
| 69 | RORA | 380164 5220215 | 15262 | -0.409 | -0.0541 | No | ||
| 70 | GPR156 | 6350609 6900079 | 15349 | -0.445 | -0.0524 | No | ||
| 71 | MDK | 3840020 | 15354 | -0.449 | -0.0462 | No | ||
| 72 | ARF1 | 3840291 | 17014 | -1.740 | -0.1109 | No | ||
| 73 | SSH2 | 360056 | 17119 | -1.869 | -0.0899 | No | ||
| 74 | DGKA | 5720152 5890328 | 17126 | -1.874 | -0.0635 | No | ||
| 75 | PTMA | 5570148 | 17343 | -2.177 | -0.0441 | No | ||
| 76 | SLCO3A1 | 1050408 2370156 6110072 | 18584 | -7.908 | 0.0017 | No |