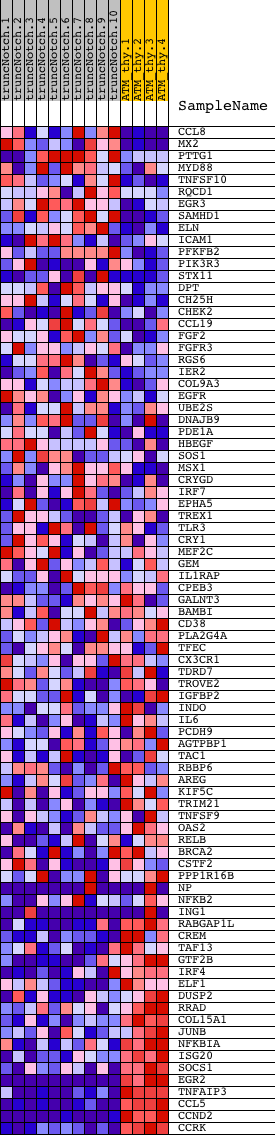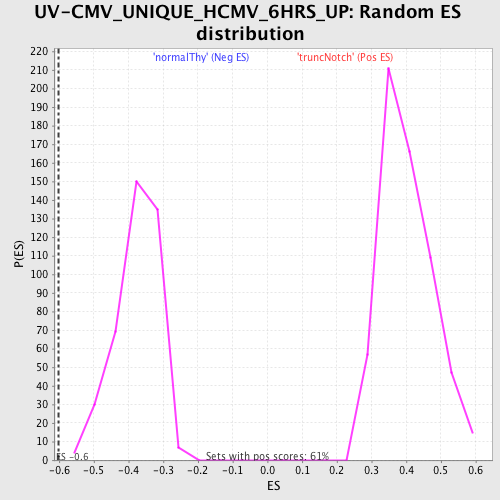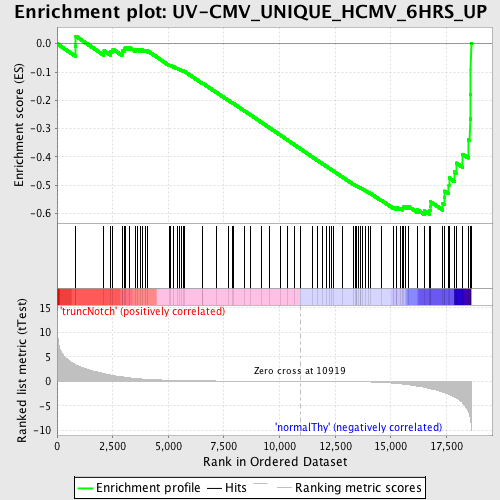
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | UV-CMV_UNIQUE_HCMV_6HRS_UP |
| Enrichment Score (ES) | -0.60302645 |
| Normalized Enrichment Score (NES) | -1.6029782 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.30056325 |
| FWER p-Value | 0.967 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CCL8 | 3870010 | 824 | 3.396 | -0.0081 | No | ||
| 2 | MX2 | 2030373 | 842 | 3.339 | 0.0267 | No | ||
| 3 | PTTG1 | 520463 | 2106 | 1.578 | -0.0245 | No | ||
| 4 | MYD88 | 2100717 | 2413 | 1.296 | -0.0272 | No | ||
| 5 | TNFSF10 | 3170300 | 2510 | 1.192 | -0.0196 | No | ||
| 6 | RQCD1 | 1850139 | 2922 | 0.938 | -0.0317 | No | ||
| 7 | EGR3 | 6940128 | 2953 | 0.918 | -0.0235 | No | ||
| 8 | SAMHD1 | 2350338 5390451 | 3024 | 0.865 | -0.0180 | No | ||
| 9 | ELN | 5080347 | 3075 | 0.832 | -0.0118 | No | ||
| 10 | ICAM1 | 6980138 | 3249 | 0.723 | -0.0134 | No | ||
| 11 | PFKFB2 | 3940538 7100059 | 3528 | 0.590 | -0.0221 | No | ||
| 12 | PIK3R3 | 1770333 | 3593 | 0.560 | -0.0195 | No | ||
| 13 | STX11 | 2470687 3710528 6590184 | 3742 | 0.511 | -0.0220 | No | ||
| 14 | DPT | 5670731 | 3830 | 0.481 | -0.0216 | No | ||
| 15 | CH25H | 4210167 | 3960 | 0.446 | -0.0238 | No | ||
| 16 | CHEK2 | 610139 1050022 | 4071 | 0.413 | -0.0253 | No | ||
| 17 | CCL19 | 5080487 | 5059 | 0.231 | -0.0761 | No | ||
| 18 | FGF2 | 3450433 | 5096 | 0.226 | -0.0756 | No | ||
| 19 | FGFR3 | 5390632 6020021 | 5253 | 0.208 | -0.0818 | No | ||
| 20 | RGS6 | 6040601 | 5390 | 0.194 | -0.0870 | No | ||
| 21 | IER2 | 2030008 | 5485 | 0.182 | -0.0902 | No | ||
| 22 | COL9A3 | 4050541 | 5586 | 0.173 | -0.0937 | No | ||
| 23 | EGFR | 4920138 6480521 | 5664 | 0.167 | -0.0961 | No | ||
| 24 | UBE2S | 1850110 | 5704 | 0.162 | -0.0964 | No | ||
| 25 | DNAJB9 | 460114 | 6522 | 0.107 | -0.1394 | No | ||
| 26 | PDE1A | 1050458 2570563 | 6541 | 0.107 | -0.1392 | No | ||
| 27 | HBEGF | 2030156 | 7179 | 0.078 | -0.1727 | No | ||
| 28 | SOS1 | 7050338 | 7718 | 0.062 | -0.2011 | No | ||
| 29 | MSX1 | 2650309 | 7864 | 0.058 | -0.2083 | No | ||
| 30 | CRYGD | 2680095 | 7936 | 0.056 | -0.2115 | No | ||
| 31 | IRF7 | 1570605 | 8402 | 0.045 | -0.2361 | No | ||
| 32 | EPHA5 | 2340671 | 8705 | 0.039 | -0.2520 | No | ||
| 33 | TREX1 | 3450040 7100692 | 9164 | 0.029 | -0.2764 | No | ||
| 34 | TLR3 | 6760451 | 9561 | 0.023 | -0.2976 | No | ||
| 35 | CRY1 | 6770039 | 10044 | 0.015 | -0.3234 | No | ||
| 36 | MEF2C | 670025 780338 | 10374 | 0.009 | -0.3411 | No | ||
| 37 | GEM | 5290082 | 10681 | 0.004 | -0.3575 | No | ||
| 38 | IL1RAP | 50546 450711 1580397 3940301 5890181 6370373 | 10939 | -0.000 | -0.3714 | No | ||
| 39 | CPEB3 | 3940164 | 11470 | -0.010 | -0.3999 | No | ||
| 40 | GALNT3 | 1850687 | 11707 | -0.014 | -0.4125 | No | ||
| 41 | BAMBI | 6650463 | 11939 | -0.019 | -0.4247 | No | ||
| 42 | CD38 | 6100056 | 12086 | -0.022 | -0.4324 | No | ||
| 43 | PLA2G4A | 6380364 | 12255 | -0.026 | -0.4412 | No | ||
| 44 | TFEC | 1170722 | 12347 | -0.028 | -0.4458 | No | ||
| 45 | CX3CR1 | 1690577 | 12441 | -0.031 | -0.4505 | No | ||
| 46 | TDRD7 | 6550520 | 12827 | -0.041 | -0.4708 | No | ||
| 47 | TROVE2 | 4850458 | 13317 | -0.061 | -0.4965 | No | ||
| 48 | IGFBP2 | 5220563 | 13396 | -0.065 | -0.5000 | No | ||
| 49 | INDO | 2680390 | 13438 | -0.067 | -0.5015 | No | ||
| 50 | IL6 | 380133 | 13465 | -0.069 | -0.5022 | No | ||
| 51 | PCDH9 | 2100136 | 13564 | -0.075 | -0.5067 | No | ||
| 52 | AGTPBP1 | 1990735 | 13628 | -0.079 | -0.5092 | No | ||
| 53 | TAC1 | 7000195 380706 | 13640 | -0.079 | -0.5090 | No | ||
| 54 | RBBP6 | 2320129 | 13725 | -0.086 | -0.5126 | No | ||
| 55 | AREG | 4920025 | 13867 | -0.096 | -0.5192 | No | ||
| 56 | KIF5C | 6370717 | 14016 | -0.110 | -0.5260 | No | ||
| 57 | TRIM21 | 3360026 | 14063 | -0.114 | -0.5272 | No | ||
| 58 | TNFSF9 | 1190408 | 14568 | -0.194 | -0.5524 | No | ||
| 59 | OAS2 | 1230408 | 15116 | -0.350 | -0.5781 | No | ||
| 60 | RELB | 1400048 | 15239 | -0.399 | -0.5804 | No | ||
| 61 | BRCA2 | 4280372 | 15256 | -0.405 | -0.5770 | No | ||
| 62 | CSTF2 | 6040463 | 15421 | -0.486 | -0.5806 | No | ||
| 63 | PPP1R16B | 3520300 | 15546 | -0.548 | -0.5814 | No | ||
| 64 | NP | 2630577 3120142 5670068 6290039 | 15561 | -0.555 | -0.5763 | No | ||
| 65 | NFKB2 | 2320670 | 15676 | -0.622 | -0.5758 | No | ||
| 66 | ING1 | 5690010 6520056 | 15794 | -0.686 | -0.5747 | No | ||
| 67 | RABGAP1L | 3190014 6860088 | 16202 | -0.961 | -0.5864 | No | ||
| 68 | CREM | 840156 6380438 6660041 6660168 | 16504 | -1.193 | -0.5899 | Yes | ||
| 69 | TAF13 | 4050692 | 16749 | -1.433 | -0.5877 | Yes | ||
| 70 | GTF2B | 6860685 | 16777 | -1.475 | -0.5733 | Yes | ||
| 71 | IRF4 | 610133 | 16782 | -1.478 | -0.5577 | Yes | ||
| 72 | ELF1 | 4780450 | 17335 | -2.163 | -0.5644 | Yes | ||
| 73 | DUSP2 | 2370184 | 17408 | -2.279 | -0.5438 | Yes | ||
| 74 | RRAD | 4280129 | 17427 | -2.311 | -0.5201 | Yes | ||
| 75 | COL15A1 | 6840113 | 17612 | -2.635 | -0.5018 | Yes | ||
| 76 | JUNB | 4230048 | 17617 | -2.644 | -0.4737 | Yes | ||
| 77 | NFKBIA | 1570152 | 17856 | -3.162 | -0.4527 | Yes | ||
| 78 | ISG20 | 1770750 | 17937 | -3.348 | -0.4212 | Yes | ||
| 79 | SOCS1 | 730139 | 18230 | -4.361 | -0.3902 | Yes | ||
| 80 | EGR2 | 3800403 | 18481 | -6.052 | -0.3389 | Yes | ||
| 81 | TNFAIP3 | 2900142 | 18558 | -7.288 | -0.2650 | Yes | ||
| 82 | CCL5 | 3710397 | 18590 | -8.151 | -0.1794 | Yes | ||
| 83 | CCND2 | 5340167 | 18602 | -8.430 | -0.0897 | Yes | ||
| 84 | CCRK | 1450161 2030500 5080112 | 18603 | -8.445 | 0.0007 | Yes |