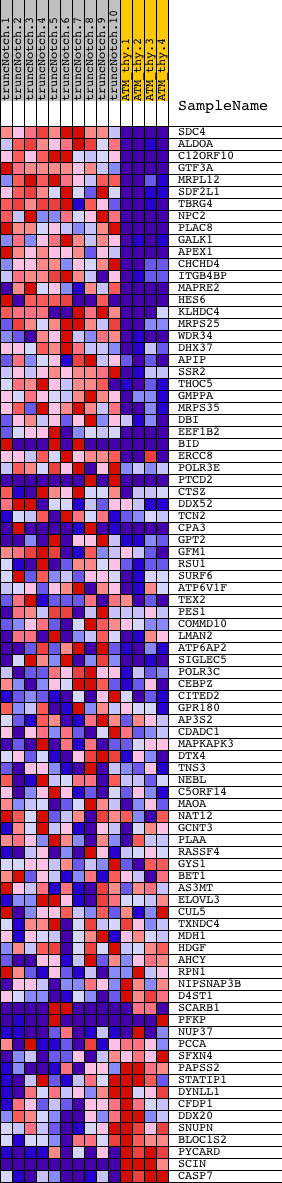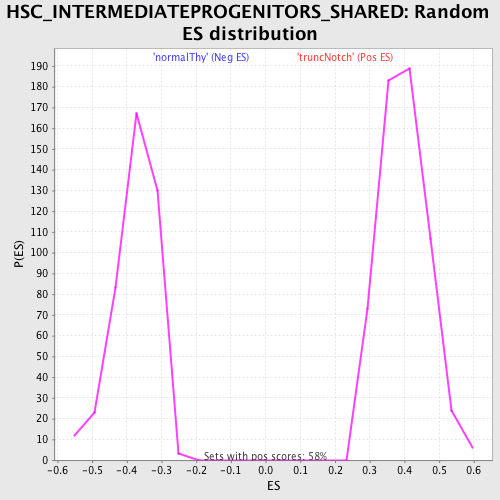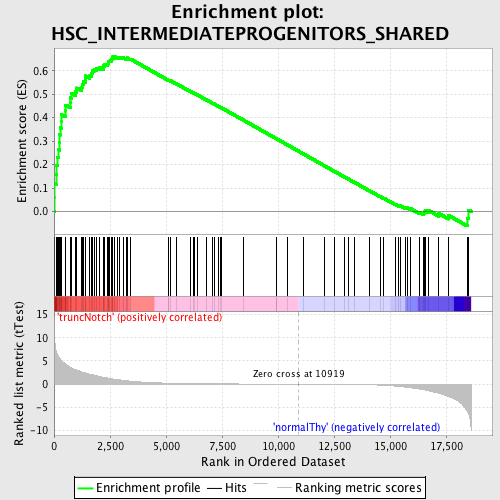
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | truncNotch |
| GeneSet | HSC_INTERMEDIATEPROGENITORS_SHARED |
| Enrichment Score (ES) | 0.6592271 |
| Normalized Enrichment Score (NES) | 1.6574783 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.038813833 |
| FWER p-Value | 0.532 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SDC4 | 6370411 | 11 | 10.535 | 0.0609 | Yes | ||
| 2 | ALDOA | 6290672 | 14 | 10.281 | 0.1209 | Yes | ||
| 3 | C12ORF10 | 4120324 5270113 | 92 | 7.107 | 0.1582 | Yes | ||
| 4 | GTF3A | 2340451 | 121 | 6.671 | 0.1957 | Yes | ||
| 5 | MRPL12 | 3120112 | 149 | 6.380 | 0.2315 | Yes | ||
| 6 | SDF2L1 | 4230446 | 188 | 5.942 | 0.2641 | Yes | ||
| 7 | TBRG4 | 3610239 6370594 | 232 | 5.682 | 0.2950 | Yes | ||
| 8 | NPC2 | 2350102 | 237 | 5.666 | 0.3279 | Yes | ||
| 9 | PLAC8 | 2900019 | 270 | 5.399 | 0.3577 | Yes | ||
| 10 | GALK1 | 840162 | 334 | 4.994 | 0.3834 | Yes | ||
| 11 | APEX1 | 3190519 | 339 | 4.957 | 0.4122 | Yes | ||
| 12 | CHCHD4 | 2680075 | 504 | 4.351 | 0.4287 | Yes | ||
| 13 | ITGB4BP | 1400541 | 510 | 4.316 | 0.4537 | Yes | ||
| 14 | MAPRE2 | 1660435 5860403 | 724 | 3.672 | 0.4636 | Yes | ||
| 15 | HES6 | 540411 6550504 | 736 | 3.640 | 0.4843 | Yes | ||
| 16 | KLHDC4 | 870288 5900092 | 767 | 3.557 | 0.5034 | Yes | ||
| 17 | MRPS25 | 610441 870128 1410368 | 956 | 3.121 | 0.5115 | Yes | ||
| 18 | WDR34 | 6100215 | 1007 | 3.032 | 0.5265 | Yes | ||
| 19 | DHX37 | 1400309 | 1238 | 2.639 | 0.5295 | Yes | ||
| 20 | APIP | 2480440 2810471 | 1264 | 2.597 | 0.5434 | Yes | ||
| 21 | SSR2 | 5390180 | 1308 | 2.522 | 0.5558 | Yes | ||
| 22 | THOC5 | 4570047 | 1388 | 2.412 | 0.5656 | Yes | ||
| 23 | GMPPA | 520450 3120497 | 1417 | 2.377 | 0.5780 | Yes | ||
| 24 | MRPS35 | 2370603 4070551 | 1598 | 2.141 | 0.5807 | Yes | ||
| 25 | DBI | 1090706 | 1676 | 2.049 | 0.5886 | Yes | ||
| 26 | EEF1B2 | 4850400 6130161 | 1697 | 2.029 | 0.5993 | Yes | ||
| 27 | BID | 4590592 6130008 | 1783 | 1.950 | 0.6061 | Yes | ||
| 28 | ERCC8 | 1240300 6450372 6590180 | 1901 | 1.813 | 0.6104 | Yes | ||
| 29 | POLR3E | 380152 | 2025 | 1.692 | 0.6136 | Yes | ||
| 30 | PTCD2 | 1230088 1400605 3170576 6200609 | 2198 | 1.495 | 0.6131 | Yes | ||
| 31 | CTSZ | 1500687 1690364 | 2218 | 1.482 | 0.6207 | Yes | ||
| 32 | DDX52 | 2970411 | 2254 | 1.455 | 0.6273 | Yes | ||
| 33 | TCN2 | 2810139 | 2376 | 1.344 | 0.6286 | Yes | ||
| 34 | CPA3 | 610242 6380102 | 2408 | 1.304 | 0.6346 | Yes | ||
| 35 | GPT2 | 1850463 | 2423 | 1.284 | 0.6413 | Yes | ||
| 36 | GFM1 | 6220711 | 2493 | 1.210 | 0.6447 | Yes | ||
| 37 | RSU1 | 4850372 | 2556 | 1.163 | 0.6481 | Yes | ||
| 38 | SURF6 | 3060451 | 2591 | 1.139 | 0.6529 | Yes | ||
| 39 | ATP6V1F | 5420082 | 2605 | 1.131 | 0.6588 | Yes | ||
| 40 | TEX2 | 1740504 2480102 3060494 | 2712 | 1.046 | 0.6592 | Yes | ||
| 41 | PES1 | 2470427 | 2843 | 0.985 | 0.6580 | No | ||
| 42 | COMMD10 | 3120451 | 2937 | 0.927 | 0.6584 | No | ||
| 43 | LMAN2 | 430154 | 3079 | 0.830 | 0.6556 | No | ||
| 44 | ATP6AP2 | 4010059 5080315 7100347 6660484 | 3251 | 0.722 | 0.6506 | No | ||
| 45 | SIGLEC5 | 5690398 | 3253 | 0.721 | 0.6547 | No | ||
| 46 | POLR3C | 6940546 | 3411 | 0.640 | 0.6500 | No | ||
| 47 | CEBPZ | 2970053 2680068 | 5118 | 0.224 | 0.5592 | No | ||
| 48 | CITED2 | 5670114 5130088 | 5122 | 0.223 | 0.5604 | No | ||
| 49 | GPR180 | 5390711 | 5217 | 0.212 | 0.5565 | No | ||
| 50 | AP3S2 | 2630411 5080053 | 5444 | 0.187 | 0.5454 | No | ||
| 51 | CDADC1 | 3710201 4120064 | 6092 | 0.130 | 0.5113 | No | ||
| 52 | MAPKAPK3 | 630520 | 6236 | 0.122 | 0.5043 | No | ||
| 53 | DTX4 | 4120433 | 6270 | 0.120 | 0.5032 | No | ||
| 54 | TNS3 | 6380242 | 6408 | 0.112 | 0.4964 | No | ||
| 55 | NEBL | 1850369 2480286 | 6821 | 0.093 | 0.4748 | No | ||
| 56 | C5ORF14 | 3780050 | 7066 | 0.083 | 0.4621 | No | ||
| 57 | MAOA | 1410039 4610324 | 7141 | 0.080 | 0.4585 | No | ||
| 58 | NAT12 | 1780092 4730370 | 7359 | 0.072 | 0.4473 | No | ||
| 59 | GCNT3 | 6290239 | 7435 | 0.069 | 0.4436 | No | ||
| 60 | PLAA | 60706 6370369 | 7454 | 0.069 | 0.4430 | No | ||
| 61 | RASSF4 | 510372 1660673 6290040 | 8432 | 0.044 | 0.3906 | No | ||
| 62 | GYS1 | 540154 | 9912 | 0.017 | 0.3108 | No | ||
| 63 | BET1 | 4010725 | 9932 | 0.016 | 0.3099 | No | ||
| 64 | AS3MT | 1570064 2350113 | 10438 | 0.008 | 0.2827 | No | ||
| 65 | ELOVL3 | 6860050 | 11133 | -0.004 | 0.2453 | No | ||
| 66 | CUL5 | 450142 | 12053 | -0.021 | 0.1958 | No | ||
| 67 | TXNDC4 | 450537 | 12518 | -0.033 | 0.1710 | No | ||
| 68 | MDH1 | 6660358 6760731 | 12975 | -0.046 | 0.1466 | No | ||
| 69 | HDGF | 5130114 5420066 6590152 | 13119 | -0.052 | 0.1392 | No | ||
| 70 | AHCY | 4230605 | 13391 | -0.065 | 0.1250 | No | ||
| 71 | RPN1 | 1240113 | 14061 | -0.114 | 0.0895 | No | ||
| 72 | NIPSNAP3B | 630672 3710091 4540288 4780576 | 14567 | -0.193 | 0.0634 | No | ||
| 73 | D4ST1 | 1780671 | 14725 | -0.231 | 0.0563 | No | ||
| 74 | SCARB1 | 510441 6770047 | 15225 | -0.393 | 0.0316 | No | ||
| 75 | PFKP | 70138 6760040 1170278 | 15394 | -0.474 | 0.0253 | No | ||
| 76 | NUP37 | 2370097 6370435 6380008 | 15445 | -0.499 | 0.0255 | No | ||
| 77 | PCCA | 3390400 | 15690 | -0.634 | 0.0161 | No | ||
| 78 | SFXN4 | 60750 4560059 | 15768 | -0.671 | 0.0158 | No | ||
| 79 | PAPSS2 | 870113 | 15898 | -0.746 | 0.0132 | No | ||
| 80 | STATIP1 | 4480022 6940097 | 16296 | -1.003 | -0.0023 | No | ||
| 81 | DYNLL1 | 5570113 | 16499 | -1.189 | -0.0063 | No | ||
| 82 | CFDP1 | 7000112 | 16547 | -1.231 | -0.0016 | No | ||
| 83 | DDX20 | 3290348 | 16567 | -1.249 | 0.0046 | No | ||
| 84 | SNUPN | 5050131 | 16729 | -1.410 | 0.0042 | No | ||
| 85 | BLOC1S2 | 1240215 | 17173 | -1.931 | -0.0085 | No | ||
| 86 | PYCARD | 2100750 | 17607 | -2.625 | -0.0165 | No | ||
| 87 | SCIN | 5420180 | 18442 | -5.806 | -0.0276 | No | ||
| 88 | CASP7 | 4210156 | 18505 | -6.324 | 0.0060 | No |