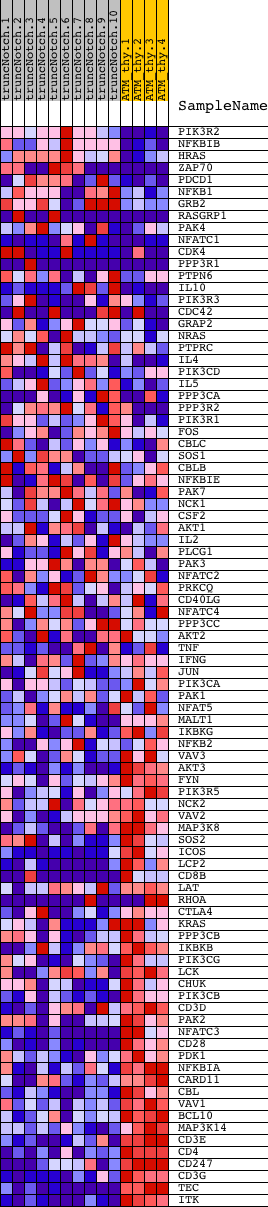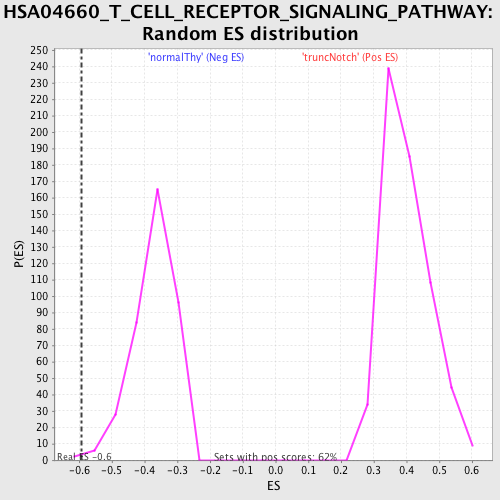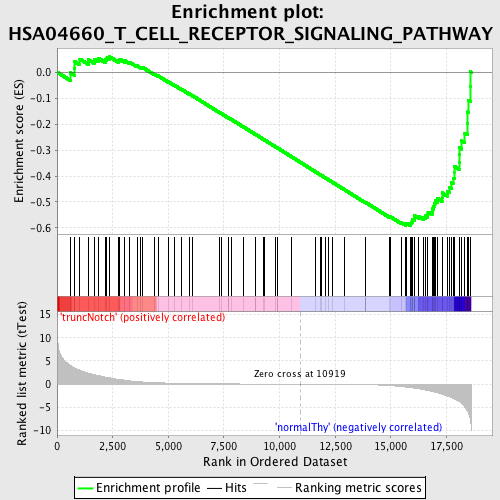
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | HSA04660_T_CELL_RECEPTOR_SIGNALING_PATHWAY |
| Enrichment Score (ES) | -0.5924187 |
| Normalized Enrichment Score (NES) | -1.5852388 |
| Nominal p-value | 0.005249344 |
| FDR q-value | 0.36513525 |
| FWER p-Value | 0.989 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PIK3R2 | 1660193 | 606 | 3.975 | -0.0026 | No | ||
| 2 | NFKBIB | 2260167 | 770 | 3.547 | 0.0154 | No | ||
| 3 | HRAS | 1980551 | 798 | 3.458 | 0.0401 | No | ||
| 4 | ZAP70 | 1410494 2260504 | 1024 | 2.988 | 0.0505 | No | ||
| 5 | PDCD1 | 4120082 | 1409 | 2.385 | 0.0478 | No | ||
| 6 | NFKB1 | 5420358 | 1663 | 2.059 | 0.0498 | No | ||
| 7 | GRB2 | 6650398 | 1856 | 1.865 | 0.0535 | No | ||
| 8 | RASGRP1 | 1050148 4050632 6510671 | 2163 | 1.527 | 0.0485 | No | ||
| 9 | PAK4 | 1690152 | 2241 | 1.462 | 0.0554 | No | ||
| 10 | NFATC1 | 510400 2320348 4050600 6180161 6290136 6620086 | 2365 | 1.357 | 0.0590 | No | ||
| 11 | CDK4 | 540075 4540600 | 2750 | 1.023 | 0.0461 | No | ||
| 12 | PPP3R1 | 1190041 1570487 | 2809 | 0.997 | 0.0505 | No | ||
| 13 | PTPN6 | 4230128 | 3027 | 0.864 | 0.0453 | No | ||
| 14 | IL10 | 2340685 2640541 2850403 6590286 | 3254 | 0.720 | 0.0385 | No | ||
| 15 | PIK3R3 | 1770333 | 3593 | 0.560 | 0.0245 | No | ||
| 16 | CDC42 | 1240168 3440278 4480519 5290162 | 3752 | 0.508 | 0.0198 | No | ||
| 17 | GRAP2 | 7100441 1410647 | 3859 | 0.474 | 0.0177 | No | ||
| 18 | NRAS | 6900577 | 4377 | 0.338 | -0.0076 | No | ||
| 19 | PTPRC | 130402 5290148 | 4545 | 0.309 | -0.0143 | No | ||
| 20 | IL4 | 6020537 | 4994 | 0.240 | -0.0367 | No | ||
| 21 | PIK3CD | 3060546 | 5286 | 0.205 | -0.0509 | No | ||
| 22 | IL5 | 6370364 1050193 | 5609 | 0.171 | -0.0669 | No | ||
| 23 | PPP3CA | 4760332 6760092 | 5932 | 0.142 | -0.0832 | No | ||
| 24 | PPP3R2 | 7000154 | 6096 | 0.130 | -0.0911 | No | ||
| 25 | PIK3R1 | 4730671 | 7309 | 0.074 | -0.1559 | No | ||
| 26 | FOS | 1850315 | 7375 | 0.071 | -0.1589 | No | ||
| 27 | CBLC | 50152 | 7689 | 0.063 | -0.1753 | No | ||
| 28 | SOS1 | 7050338 | 7718 | 0.062 | -0.1764 | No | ||
| 29 | CBLB | 3520465 4670348 | 7847 | 0.058 | -0.1828 | No | ||
| 30 | NFKBIE | 580390 2190086 | 8369 | 0.045 | -0.2106 | No | ||
| 31 | PAK7 | 3830164 | 8919 | 0.034 | -0.2400 | No | ||
| 32 | NCK1 | 6200575 6510050 | 8925 | 0.033 | -0.2400 | No | ||
| 33 | CSF2 | 1660593 | 9269 | 0.028 | -0.2583 | No | ||
| 34 | AKT1 | 5290746 | 9328 | 0.027 | -0.2612 | No | ||
| 35 | IL2 | 1770725 | 9836 | 0.018 | -0.2885 | No | ||
| 36 | PLCG1 | 6020369 | 9889 | 0.017 | -0.2911 | No | ||
| 37 | PAK3 | 4210136 | 9921 | 0.016 | -0.2927 | No | ||
| 38 | NFATC2 | 70450 540097 | 10515 | 0.007 | -0.3246 | No | ||
| 39 | PRKCQ | 2260170 3870193 | 11611 | -0.012 | -0.3837 | No | ||
| 40 | CD40LG | 3120270 | 11851 | -0.017 | -0.3964 | No | ||
| 41 | NFATC4 | 2470735 | 11895 | -0.018 | -0.3986 | No | ||
| 42 | PPP3CC | 2450139 | 12057 | -0.021 | -0.4071 | No | ||
| 43 | AKT2 | 3850541 3870519 | 12186 | -0.025 | -0.4139 | No | ||
| 44 | TNF | 6650603 | 12392 | -0.029 | -0.4247 | No | ||
| 45 | IFNG | 5670592 | 12899 | -0.044 | -0.4517 | No | ||
| 46 | JUN | 840170 | 13860 | -0.095 | -0.5028 | No | ||
| 47 | PIK3CA | 6220129 | 13874 | -0.096 | -0.5028 | No | ||
| 48 | PAK1 | 4540315 | 13880 | -0.097 | -0.5023 | No | ||
| 49 | NFAT5 | 2510411 5890195 6550152 | 14931 | -0.293 | -0.5567 | No | ||
| 50 | MALT1 | 4670292 | 14981 | -0.308 | -0.5571 | No | ||
| 51 | IKBKG | 3450092 3840377 6590592 | 15489 | -0.523 | -0.5805 | No | ||
| 52 | NFKB2 | 2320670 | 15676 | -0.622 | -0.5858 | No | ||
| 53 | VAV3 | 1050731 2450242 2680653 | 15709 | -0.646 | -0.5826 | No | ||
| 54 | AKT3 | 1580270 3290278 | 15891 | -0.742 | -0.5868 | Yes | ||
| 55 | FYN | 2100468 4760520 4850687 | 15896 | -0.745 | -0.5814 | Yes | ||
| 56 | PIK3R5 | 1090056 | 15924 | -0.763 | -0.5771 | Yes | ||
| 57 | NCK2 | 2510010 | 15978 | -0.794 | -0.5739 | Yes | ||
| 58 | VAV2 | 3610725 5890717 | 15989 | -0.803 | -0.5684 | Yes | ||
| 59 | MAP3K8 | 2940286 | 16052 | -0.854 | -0.5653 | Yes | ||
| 60 | SOS2 | 2060722 | 16061 | -0.863 | -0.5592 | Yes | ||
| 61 | ICOS | 1190048 | 16064 | -0.868 | -0.5527 | Yes | ||
| 62 | LCP2 | 2680066 6650707 | 16263 | -0.994 | -0.5559 | Yes | ||
| 63 | CD8B | 1940411 2570035 | 16471 | -1.157 | -0.5583 | Yes | ||
| 64 | LAT | 3170025 | 16561 | -1.244 | -0.5537 | Yes | ||
| 65 | RHOA | 580142 5900131 5340450 | 16657 | -1.337 | -0.5487 | Yes | ||
| 66 | CTLA4 | 6590537 | 16670 | -1.347 | -0.5392 | Yes | ||
| 67 | KRAS | 2060170 | 16854 | -1.572 | -0.5372 | Yes | ||
| 68 | PPP3CB | 6020156 | 16865 | -1.582 | -0.5258 | Yes | ||
| 69 | IKBKB | 6840072 | 16924 | -1.659 | -0.5163 | Yes | ||
| 70 | PIK3CG | 5890110 | 16947 | -1.678 | -0.5048 | Yes | ||
| 71 | LCK | 3360142 | 17003 | -1.730 | -0.4947 | Yes | ||
| 72 | CHUK | 7050736 | 17108 | -1.856 | -0.4863 | Yes | ||
| 73 | PIK3CB | 3800600 | 17306 | -2.122 | -0.4809 | Yes | ||
| 74 | CD3D | 2810739 | 17311 | -2.126 | -0.4650 | Yes | ||
| 75 | PAK2 | 360438 7050068 | 17539 | -2.495 | -0.4584 | Yes | ||
| 76 | NFATC3 | 5050520 | 17626 | -2.654 | -0.4430 | Yes | ||
| 77 | CD28 | 1400739 4210093 | 17714 | -2.847 | -0.4261 | Yes | ||
| 78 | PDK1 | 5290427 | 17798 | -3.019 | -0.4078 | Yes | ||
| 79 | NFKBIA | 1570152 | 17856 | -3.162 | -0.3869 | Yes | ||
| 80 | CARD11 | 70338 | 17860 | -3.175 | -0.3631 | Yes | ||
| 81 | CBL | 6380068 | 18068 | -3.711 | -0.3462 | Yes | ||
| 82 | VAV1 | 6020487 | 18073 | -3.720 | -0.3183 | Yes | ||
| 83 | BCL10 | 2360397 | 18081 | -3.744 | -0.2903 | Yes | ||
| 84 | MAP3K14 | 5890435 | 18190 | -4.158 | -0.2647 | Yes | ||
| 85 | CD3E | 3800056 | 18329 | -4.901 | -0.2351 | Yes | ||
| 86 | CD4 | 1090010 | 18448 | -5.828 | -0.1974 | Yes | ||
| 87 | CD247 | 3800725 5720136 | 18455 | -5.882 | -0.1532 | Yes | ||
| 88 | CD3G | 2680288 | 18503 | -6.298 | -0.1081 | Yes | ||
| 89 | TEC | 1400576 | 18565 | -7.453 | -0.0551 | Yes | ||
| 90 | ITK | 2230592 | 18571 | -7.638 | 0.0024 | Yes |