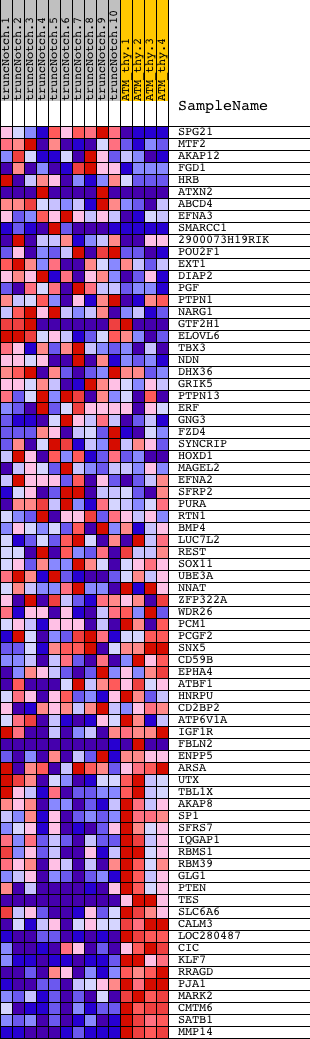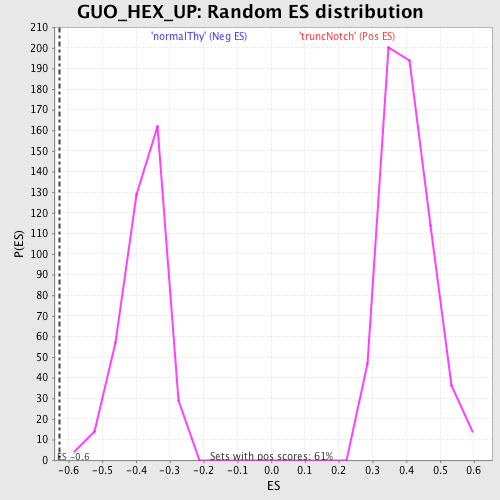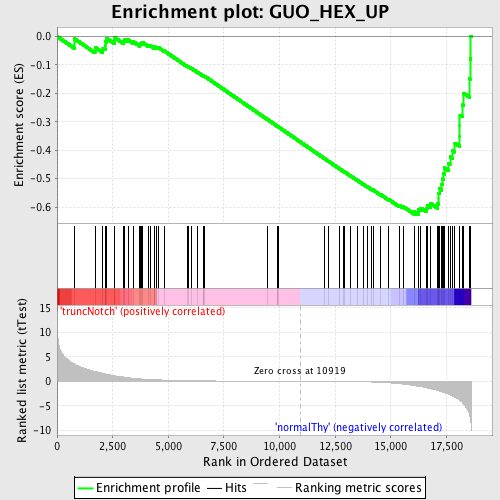
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | GUO_HEX_UP |
| Enrichment Score (ES) | -0.62590164 |
| Normalized Enrichment Score (NES) | -1.6573889 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.2331502 |
| FWER p-Value | 0.775 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SPG21 | 2060619 | 785 | 3.510 | -0.0084 | No | ||
| 2 | MTF2 | 50333 | 1707 | 2.019 | -0.0385 | No | ||
| 3 | AKAP12 | 1450739 | 2060 | 1.646 | -0.0416 | No | ||
| 4 | FGD1 | 4780021 | 2176 | 1.514 | -0.0331 | No | ||
| 5 | HRB | 1660397 6110398 | 2180 | 1.509 | -0.0187 | No | ||
| 6 | ATXN2 | 3940017 4810242 | 2222 | 1.479 | -0.0066 | No | ||
| 7 | ABCD4 | 1450164 | 2567 | 1.152 | -0.0140 | No | ||
| 8 | EFNA3 | 6370390 | 2598 | 1.134 | -0.0046 | No | ||
| 9 | SMARCC1 | 5080019 7100047 | 2987 | 0.894 | -0.0169 | No | ||
| 10 | 2900073H19RIK | 4920369 | 3041 | 0.855 | -0.0115 | No | ||
| 11 | POU2F1 | 70577 430373 4850324 5910056 | 3188 | 0.760 | -0.0120 | No | ||
| 12 | EXT1 | 4540603 | 3430 | 0.631 | -0.0189 | No | ||
| 13 | DIAP2 | 3060070 | 3719 | 0.518 | -0.0294 | No | ||
| 14 | PGF | 4810593 | 3740 | 0.512 | -0.0255 | No | ||
| 15 | PTPN1 | 2650056 | 3786 | 0.499 | -0.0231 | No | ||
| 16 | NARG1 | 5910563 6350095 | 3824 | 0.482 | -0.0205 | No | ||
| 17 | GTF2H1 | 2570746 2650148 | 4099 | 0.405 | -0.0313 | No | ||
| 18 | ELOVL6 | 5340746 | 4182 | 0.386 | -0.0320 | No | ||
| 19 | TBX3 | 2570672 | 4388 | 0.336 | -0.0398 | No | ||
| 20 | NDN | 5670075 | 4398 | 0.335 | -0.0370 | No | ||
| 21 | DHX36 | 2470465 | 4462 | 0.324 | -0.0373 | No | ||
| 22 | GRIK5 | 1710196 3710717 | 4540 | 0.310 | -0.0385 | No | ||
| 23 | PTPN13 | 2060681 | 4822 | 0.262 | -0.0511 | No | ||
| 24 | ERF | 110551 1770278 | 5856 | 0.147 | -0.1054 | No | ||
| 25 | GNG3 | 6980397 | 5918 | 0.143 | -0.1073 | No | ||
| 26 | FZD4 | 1580520 | 6048 | 0.133 | -0.1130 | No | ||
| 27 | SYNCRIP | 1690195 3140113 4670279 | 6326 | 0.117 | -0.1268 | No | ||
| 28 | HOXD1 | 7000458 | 6586 | 0.104 | -0.1397 | No | ||
| 29 | MAGEL2 | 7050619 | 6587 | 0.104 | -0.1387 | No | ||
| 30 | EFNA2 | 460193 | 6630 | 0.102 | -0.1400 | No | ||
| 31 | SFRP2 | 4850097 | 9442 | 0.025 | -0.2914 | No | ||
| 32 | PURA | 3870156 | 9894 | 0.017 | -0.3155 | No | ||
| 33 | RTN1 | 4150452 4850176 | 9911 | 0.017 | -0.3162 | No | ||
| 34 | BMP4 | 380113 | 9938 | 0.016 | -0.3175 | No | ||
| 35 | LUC7L2 | 1850402 6350088 | 12024 | -0.021 | -0.4297 | No | ||
| 36 | REST | 1500471 | 12219 | -0.025 | -0.4400 | No | ||
| 37 | SOX11 | 610279 | 12687 | -0.037 | -0.4648 | No | ||
| 38 | UBE3A | 1240152 2690438 5860609 | 12866 | -0.043 | -0.4740 | No | ||
| 39 | NNAT | 4920053 5130253 | 12904 | -0.044 | -0.4755 | No | ||
| 40 | ZFP322A | 4920014 | 13169 | -0.054 | -0.4893 | No | ||
| 41 | WDR26 | 1090138 | 13514 | -0.071 | -0.5071 | No | ||
| 42 | PCM1 | 1940070 3940707 | 13782 | -0.090 | -0.5206 | No | ||
| 43 | PCGF2 | 6370347 | 13946 | -0.103 | -0.5284 | No | ||
| 44 | SNX5 | 1570021 6860609 | 14144 | -0.123 | -0.5379 | No | ||
| 45 | CD59B | 610112 | 14199 | -0.128 | -0.5396 | No | ||
| 46 | EPHA4 | 460750 | 14537 | -0.187 | -0.5559 | No | ||
| 47 | ATBF1 | 1240131 7040465 | 14897 | -0.284 | -0.5725 | No | ||
| 48 | HNRPU | 6520736 | 15375 | -0.464 | -0.5938 | No | ||
| 49 | CD2BP2 | 1580301 | 15550 | -0.550 | -0.5978 | No | ||
| 50 | ATP6V1A | 6590242 | 16058 | -0.861 | -0.6168 | Yes | ||
| 51 | IGF1R | 3360494 | 16227 | -0.973 | -0.6165 | Yes | ||
| 52 | FBLN2 | 730601 3440215 | 16235 | -0.979 | -0.6074 | Yes | ||
| 53 | ENPP5 | 1780338 | 16345 | -1.040 | -0.6032 | Yes | ||
| 54 | ARSA | 5570717 | 16591 | -1.273 | -0.6041 | Yes | ||
| 55 | UTX | 2900017 6770452 | 16636 | -1.322 | -0.5937 | Yes | ||
| 56 | TBL1X | 6400524 | 16796 | -1.501 | -0.5877 | Yes | ||
| 57 | AKAP8 | 4210026 | 17097 | -1.838 | -0.5861 | Yes | ||
| 58 | SP1 | 6590017 | 17158 | -1.914 | -0.5708 | Yes | ||
| 59 | SFRS7 | 2760408 | 17159 | -1.914 | -0.5523 | Yes | ||
| 60 | IQGAP1 | 6660494 | 17182 | -1.938 | -0.5347 | Yes | ||
| 61 | RBMS1 | 6400014 | 17294 | -2.106 | -0.5203 | Yes | ||
| 62 | RBM39 | 4230458 6450280 | 17321 | -2.138 | -0.5010 | Yes | ||
| 63 | GLG1 | 3130021 | 17379 | -2.238 | -0.4825 | Yes | ||
| 64 | PTEN | 3390064 | 17414 | -2.288 | -0.4621 | Yes | ||
| 65 | TES | 2680184 3840736 | 17605 | -2.622 | -0.4470 | Yes | ||
| 66 | SLC6A6 | 6100300 | 17662 | -2.751 | -0.4234 | Yes | ||
| 67 | CALM3 | 3390288 | 17770 | -2.963 | -0.4005 | Yes | ||
| 68 | LOC280487 | 5910112 | 17881 | -3.225 | -0.3752 | Yes | ||
| 69 | CIC | 6370161 | 18102 | -3.805 | -0.3502 | Yes | ||
| 70 | KLF7 | 50390 | 18105 | -3.812 | -0.3135 | Yes | ||
| 71 | RRAGD | 670075 | 18107 | -3.814 | -0.2766 | Yes | ||
| 72 | PJA1 | 2970600 | 18211 | -4.262 | -0.2409 | Yes | ||
| 73 | MARK2 | 7210608 | 18271 | -4.582 | -0.1997 | Yes | ||
| 74 | CMTM6 | 6590010 | 18531 | -6.754 | -0.1483 | Yes | ||
| 75 | SATB1 | 5670154 | 18564 | -7.416 | -0.0783 | Yes | ||
| 76 | MMP14 | 670079 | 18598 | -8.374 | 0.0010 | Yes |