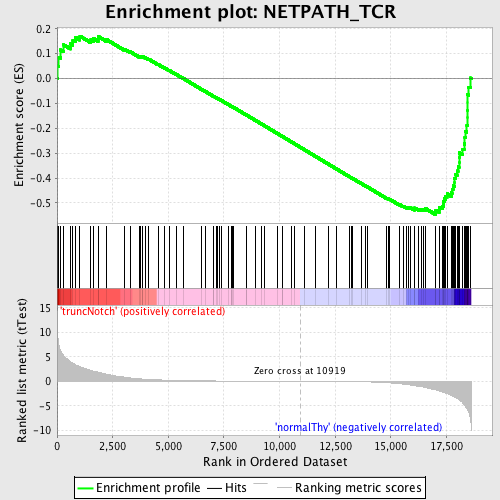
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | NETPATH_TCR |
| Enrichment Score (ES) | -0.54631925 |
| Normalized Enrichment Score (NES) | -1.5086423 |
| Nominal p-value | 0.0029239766 |
| FDR q-value | 0.39733523 |
| FWER p-Value | 0.997 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CEBPB | 2970019 | 19 | 9.933 | 0.0514 | No | ||
| 2 | UNC119 | 360372 | 82 | 7.253 | 0.0864 | No | ||
| 3 | STAT5A | 2680458 | 160 | 6.254 | 0.1152 | No | ||
| 4 | DTX1 | 5900372 | 296 | 5.224 | 0.1355 | No | ||
| 5 | PIK3R2 | 1660193 | 606 | 3.975 | 0.1398 | No | ||
| 6 | DBNL | 7100088 | 709 | 3.701 | 0.1538 | No | ||
| 7 | JAK3 | 70347 3290008 | 822 | 3.400 | 0.1657 | No | ||
| 8 | ZAP70 | 1410494 2260504 | 1024 | 2.988 | 0.1706 | No | ||
| 9 | NCL | 2360463 4540279 | 1519 | 2.258 | 0.1559 | No | ||
| 10 | HOMER3 | 3830279 | 1628 | 2.104 | 0.1611 | No | ||
| 11 | PTK2 | 1780148 | 1842 | 1.887 | 0.1596 | No | ||
| 12 | GRB2 | 6650398 | 1856 | 1.865 | 0.1687 | No | ||
| 13 | YWHAQ | 6760524 | 2230 | 1.476 | 0.1564 | No | ||
| 14 | PTPN6 | 4230128 | 3027 | 0.864 | 0.1179 | No | ||
| 15 | MAP4K1 | 4060692 | 3281 | 0.709 | 0.1080 | No | ||
| 16 | ABL1 | 1050593 2030050 4010114 | 3714 | 0.520 | 0.0874 | No | ||
| 17 | CDC42 | 1240168 3440278 4480519 5290162 | 3752 | 0.508 | 0.0881 | No | ||
| 18 | SHB | 4920494 | 3849 | 0.476 | 0.0854 | No | ||
| 19 | GRAP2 | 7100441 1410647 | 3859 | 0.474 | 0.0875 | No | ||
| 20 | PAG1 | 5910397 | 3950 | 0.448 | 0.0850 | No | ||
| 21 | DLG1 | 2630091 6020286 | 4123 | 0.399 | 0.0778 | No | ||
| 22 | PTPRC | 130402 5290148 | 4545 | 0.309 | 0.0567 | No | ||
| 23 | ENAH | 1690292 5700300 | 4829 | 0.261 | 0.0428 | No | ||
| 24 | STK39 | 50692 | 5057 | 0.232 | 0.0317 | No | ||
| 25 | DUSP3 | 1240129 6840215 | 5358 | 0.197 | 0.0166 | No | ||
| 26 | SH3BP2 | 610131 | 5676 | 0.166 | 0.0003 | No | ||
| 27 | CD2AP | 1940369 | 6490 | 0.109 | -0.0430 | No | ||
| 28 | TUBB2A | 6940435 | 6651 | 0.100 | -0.0512 | No | ||
| 29 | TXK | 2320338 | 7020 | 0.085 | -0.0706 | No | ||
| 30 | VASP | 7050500 | 7044 | 0.083 | -0.0714 | No | ||
| 31 | PTPRH | 3870070 | 7145 | 0.080 | -0.0764 | No | ||
| 32 | SH2D3C | 3060014 | 7196 | 0.077 | -0.0787 | No | ||
| 33 | PIK3R1 | 4730671 | 7309 | 0.074 | -0.0843 | No | ||
| 34 | FOS | 1850315 | 7375 | 0.071 | -0.0875 | No | ||
| 35 | SOS1 | 7050338 | 7718 | 0.062 | -0.1056 | No | ||
| 36 | ARHGEF6 | 4780164 6380450 | 7844 | 0.058 | -0.1121 | No | ||
| 37 | CBLB | 3520465 4670348 | 7847 | 0.058 | -0.1119 | No | ||
| 38 | PTPRJ | 4010707 | 7875 | 0.057 | -0.1130 | No | ||
| 39 | LYN | 6040600 | 7944 | 0.056 | -0.1164 | No | ||
| 40 | PTPN22 | 1690647 7100239 | 8491 | 0.043 | -0.1457 | No | ||
| 41 | PTPN11 | 2230100 2470180 6100528 | 8521 | 0.042 | -0.1470 | No | ||
| 42 | NCK1 | 6200575 6510050 | 8925 | 0.033 | -0.1686 | No | ||
| 43 | SKAP2 | 6760500 | 9165 | 0.029 | -0.1814 | No | ||
| 44 | AKT1 | 5290746 | 9328 | 0.027 | -0.1900 | No | ||
| 45 | PLCG1 | 6020369 | 9889 | 0.017 | -0.2201 | No | ||
| 46 | PTPN3 | 870369 | 10125 | 0.013 | -0.2327 | No | ||
| 47 | GAB2 | 1410280 2340520 4280040 | 10136 | 0.013 | -0.2332 | No | ||
| 48 | NFATC2 | 70450 540097 | 10515 | 0.007 | -0.2536 | No | ||
| 49 | INSL3 | 4150092 | 10652 | 0.005 | -0.2609 | No | ||
| 50 | RAP1A | 1090025 | 11113 | -0.003 | -0.2858 | No | ||
| 51 | PRKCQ | 2260170 3870193 | 11611 | -0.012 | -0.3125 | No | ||
| 52 | ACP1 | 1990435 | 12183 | -0.024 | -0.3433 | No | ||
| 53 | RAPGEF1 | 60040 4590670 | 12555 | -0.033 | -0.3631 | No | ||
| 54 | ARHGEF7 | 1690441 | 13151 | -0.053 | -0.3950 | No | ||
| 55 | SRC | 580132 | 13241 | -0.057 | -0.3995 | No | ||
| 56 | GRAP | 1770195 | 13272 | -0.059 | -0.4008 | No | ||
| 57 | SYK | 6940133 | 13698 | -0.084 | -0.4233 | No | ||
| 58 | JUN | 840170 | 13860 | -0.095 | -0.4315 | No | ||
| 59 | STAT5B | 6200026 | 13964 | -0.105 | -0.4365 | No | ||
| 60 | CREB1 | 1500717 2230358 3610600 6550601 | 14820 | -0.260 | -0.4813 | No | ||
| 61 | ARHGDIB | 4730433 | 14873 | -0.277 | -0.4827 | No | ||
| 62 | PTPN12 | 2030309 6020725 6130746 6290609 | 14946 | -0.297 | -0.4850 | No | ||
| 63 | SHC1 | 2900731 3170504 6520537 | 15383 | -0.470 | -0.5061 | No | ||
| 64 | MAP2K1 | 840739 | 15572 | -0.564 | -0.5132 | No | ||
| 65 | VAV3 | 1050731 2450242 2680653 | 15709 | -0.646 | -0.5172 | No | ||
| 66 | MAPK1 | 3190193 6200253 | 15779 | -0.675 | -0.5173 | No | ||
| 67 | FYN | 2100468 4760520 4850687 | 15896 | -0.745 | -0.5197 | No | ||
| 68 | SOS2 | 2060722 | 16061 | -0.863 | -0.5240 | No | ||
| 69 | FYB | 1690440 | 16076 | -0.876 | -0.5201 | No | ||
| 70 | LCP2 | 2680066 6650707 | 16263 | -0.994 | -0.5249 | No | ||
| 71 | RIPK2 | 5050072 6290632 | 16369 | -1.063 | -0.5250 | No | ||
| 72 | CRK | 1230162 4780128 | 16481 | -1.169 | -0.5248 | No | ||
| 73 | LAT | 3170025 | 16561 | -1.244 | -0.5225 | No | ||
| 74 | LCK | 3360142 | 17003 | -1.730 | -0.5372 | Yes | ||
| 75 | WIPF1 | 1170577 | 17020 | -1.746 | -0.5288 | Yes | ||
| 76 | ABI1 | 2850152 | 17170 | -1.923 | -0.5267 | Yes | ||
| 77 | CREBBP | 5690035 7040050 | 17180 | -1.936 | -0.5170 | Yes | ||
| 78 | CD3D | 2810739 | 17311 | -2.126 | -0.5128 | Yes | ||
| 79 | GIT2 | 6040039 6770181 | 17362 | -2.201 | -0.5039 | Yes | ||
| 80 | EVL | 1740113 | 17376 | -2.229 | -0.4928 | Yes | ||
| 81 | KHDRBS1 | 1240403 6040040 | 17415 | -2.292 | -0.4828 | Yes | ||
| 82 | WAS | 5270193 | 17462 | -2.355 | -0.4728 | Yes | ||
| 83 | TUBA4A | 6900279 | 17527 | -2.473 | -0.4632 | Yes | ||
| 84 | DOCK2 | 2940040 | 17729 | -2.879 | -0.4589 | Yes | ||
| 85 | SLA | 2000070 | 17755 | -2.940 | -0.4447 | Yes | ||
| 86 | WASF2 | 4850592 | 17800 | -3.026 | -0.4311 | Yes | ||
| 87 | CARD11 | 70338 | 17860 | -3.175 | -0.4175 | Yes | ||
| 88 | SH2D2A | 2970594 | 17865 | -3.186 | -0.4009 | Yes | ||
| 89 | PSTPIP1 | 3190156 | 17915 | -3.297 | -0.3862 | Yes | ||
| 90 | DNM2 | 50176 3170279 | 17981 | -3.481 | -0.3713 | Yes | ||
| 91 | RASA1 | 1240315 | 18023 | -3.576 | -0.3546 | Yes | ||
| 92 | CBL | 6380068 | 18068 | -3.711 | -0.3374 | Yes | ||
| 93 | VAV1 | 6020487 | 18073 | -3.720 | -0.3180 | Yes | ||
| 94 | BCL10 | 2360397 | 18081 | -3.744 | -0.2986 | Yes | ||
| 95 | NEDD9 | 1740373 4230053 | 18235 | -4.404 | -0.2836 | Yes | ||
| 96 | DEF6 | 840593 | 18296 | -4.710 | -0.2620 | Yes | ||
| 97 | CD3E | 3800056 | 18329 | -4.901 | -0.2379 | Yes | ||
| 98 | PTK2B | 4730411 | 18348 | -5.010 | -0.2124 | Yes | ||
| 99 | CRKL | 4050427 | 18419 | -5.557 | -0.1868 | Yes | ||
| 100 | CISH | 840315 | 18446 | -5.822 | -0.1575 | Yes | ||
| 101 | CD4 | 1090010 | 18448 | -5.828 | -0.1268 | Yes | ||
| 102 | CD5 | 6590338 | 18454 | -5.879 | -0.0960 | Yes | ||
| 103 | CD247 | 3800725 5720136 | 18455 | -5.882 | -0.0650 | Yes | ||
| 104 | CD3G | 2680288 | 18503 | -6.298 | -0.0343 | Yes | ||
| 105 | ITK | 2230592 | 18571 | -7.638 | 0.0024 | Yes |

