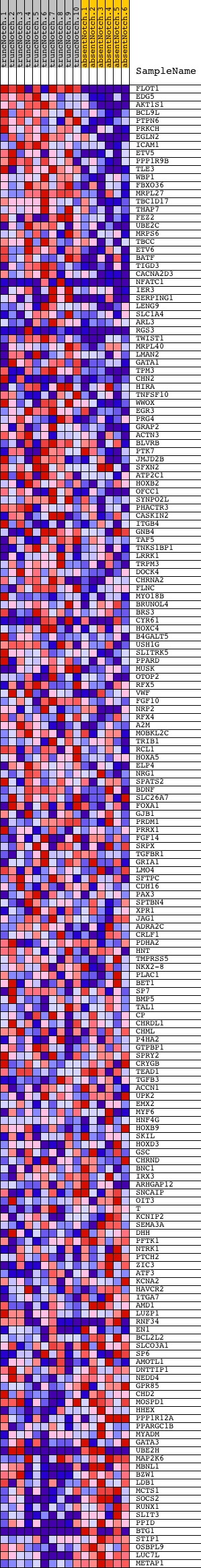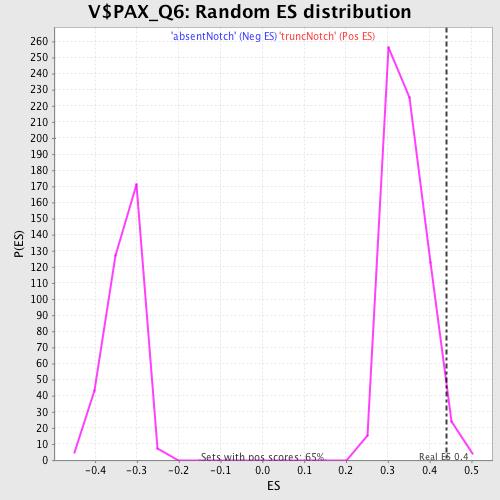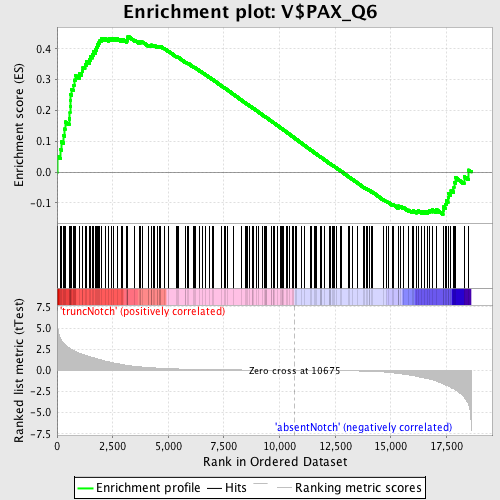
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #truncNotch_versus_absentNotch |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#truncNotch_versus_absentNotch |
| Upregulated in class | truncNotch |
| GeneSet | V$PAX_Q6 |
| Enrichment Score (ES) | 0.44144616 |
| Normalized Enrichment Score (NES) | 1.2845558 |
| Nominal p-value | 0.02782071 |
| FDR q-value | 0.9657697 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | FLOT1 | 1660575 1690040 | 2 | 6.562 | 0.0514 | Yes | ||
| 2 | EDG5 | 2640300 | 150 | 3.760 | 0.0729 | Yes | ||
| 3 | AKT1S1 | 1660048 | 200 | 3.525 | 0.0980 | Yes | ||
| 4 | BCL9L | 6940053 | 298 | 3.207 | 0.1179 | Yes | ||
| 5 | PTPN6 | 4230128 | 340 | 3.085 | 0.1399 | Yes | ||
| 6 | PRKCH | 5720079 | 360 | 3.044 | 0.1627 | Yes | ||
| 7 | EGLN2 | 540086 | 543 | 2.645 | 0.1736 | Yes | ||
| 8 | ICAM1 | 6980138 | 577 | 2.609 | 0.1923 | Yes | ||
| 9 | ETV5 | 110017 | 582 | 2.603 | 0.2125 | Yes | ||
| 10 | PPP1R9B | 3130619 | 599 | 2.558 | 0.2317 | Yes | ||
| 11 | TLE3 | 580040 4730121 | 609 | 2.546 | 0.2512 | Yes | ||
| 12 | WBP1 | 6100066 | 649 | 2.495 | 0.2687 | Yes | ||
| 13 | FBXO36 | 1410086 | 740 | 2.343 | 0.2822 | Yes | ||
| 14 | MRPL27 | 2060131 | 766 | 2.304 | 0.2989 | Yes | ||
| 15 | TBC1D17 | 1780450 | 813 | 2.248 | 0.3141 | Yes | ||
| 16 | THAP7 | 6660673 | 1001 | 2.020 | 0.3198 | Yes | ||
| 17 | FEZ2 | 5910181 | 1118 | 1.902 | 0.3284 | Yes | ||
| 18 | UBE2C | 6130017 | 1159 | 1.873 | 0.3410 | Yes | ||
| 19 | MRPS6 | 870164 | 1254 | 1.772 | 0.3498 | Yes | ||
| 20 | TBCC | 4810021 | 1316 | 1.723 | 0.3600 | Yes | ||
| 21 | ETV6 | 610524 | 1466 | 1.594 | 0.3644 | Yes | ||
| 22 | BATF | 6760390 | 1501 | 1.569 | 0.3749 | Yes | ||
| 23 | TIGD3 | 3850576 | 1587 | 1.511 | 0.3821 | Yes | ||
| 24 | CACNA2D3 | 4010184 | 1642 | 1.486 | 0.3909 | Yes | ||
| 25 | NFATC1 | 510400 2320348 4050600 6180161 6290136 6620086 | 1737 | 1.420 | 0.3969 | Yes | ||
| 26 | IER3 | 5860021 | 1773 | 1.389 | 0.4059 | Yes | ||
| 27 | SERPING1 | 5550440 | 1821 | 1.332 | 0.4138 | Yes | ||
| 28 | LENG9 | 3060072 | 1877 | 1.294 | 0.4210 | Yes | ||
| 29 | SLC1A4 | 5290064 | 1923 | 1.264 | 0.4285 | Yes | ||
| 30 | ARL3 | 5690725 | 1988 | 1.217 | 0.4346 | Yes | ||
| 31 | RGS3 | 60670 540736 1340180 1500369 3390735 4010131 4610402 6380114 | 2154 | 1.101 | 0.4343 | Yes | ||
| 32 | TWIST1 | 5130619 | 2330 | 1.005 | 0.4327 | Yes | ||
| 33 | MRPL40 | 2370215 | 2424 | 0.951 | 0.4351 | Yes | ||
| 34 | LMAN2 | 430154 | 2552 | 0.876 | 0.4351 | Yes | ||
| 35 | GATA1 | 6400068 | 2702 | 0.799 | 0.4332 | Yes | ||
| 36 | TPM3 | 670600 3170296 5670167 6650471 | 2876 | 0.702 | 0.4294 | Yes | ||
| 37 | CHN2 | 870528 | 2952 | 0.671 | 0.4306 | Yes | ||
| 38 | HIRA | 4480609 5270167 | 3125 | 0.601 | 0.4260 | Yes | ||
| 39 | TNFSF10 | 3170300 | 3144 | 0.594 | 0.4296 | Yes | ||
| 40 | WWOX | 510008 870524 1090114 | 3162 | 0.588 | 0.4333 | Yes | ||
| 41 | EGR3 | 6940128 | 3176 | 0.582 | 0.4372 | Yes | ||
| 42 | PRG4 | 5860156 6650086 | 3183 | 0.581 | 0.4414 | Yes | ||
| 43 | GRAP2 | 7100441 1410647 | 3499 | 0.469 | 0.4280 | No | ||
| 44 | ACTN3 | 3140541 6480598 | 3698 | 0.418 | 0.4206 | No | ||
| 45 | BLVRB | 7040707 | 3730 | 0.410 | 0.4221 | No | ||
| 46 | PTK7 | 380315 | 3756 | 0.404 | 0.4239 | No | ||
| 47 | JMJD2B | 6350372 6400471 | 3840 | 0.387 | 0.4225 | No | ||
| 48 | SFXN2 | 2650563 3140095 3830762 3870398 | 4089 | 0.335 | 0.4116 | No | ||
| 49 | ATP2C1 | 2630446 6520253 | 4121 | 0.331 | 0.4126 | No | ||
| 50 | HOXB2 | 6450592 | 4220 | 0.314 | 0.4097 | No | ||
| 51 | OFCC1 | 2060392 | 4239 | 0.311 | 0.4112 | No | ||
| 52 | SYNPO2L | 5390719 | 4256 | 0.308 | 0.4127 | No | ||
| 53 | PHACTR3 | 3850435 5900445 | 4336 | 0.294 | 0.4108 | No | ||
| 54 | CASKIN2 | 2030113 | 4368 | 0.288 | 0.4113 | No | ||
| 55 | ITGB4 | 1740021 3840482 | 4518 | 0.265 | 0.4053 | No | ||
| 56 | GNB4 | 3390079 4230035 | 4520 | 0.265 | 0.4074 | No | ||
| 57 | TAF5 | 3450288 5890193 6860435 | 4591 | 0.254 | 0.4056 | No | ||
| 58 | TNKS1BP1 | 7040184 | 4620 | 0.250 | 0.4060 | No | ||
| 59 | LRRK1 | 2810609 | 4626 | 0.250 | 0.4077 | No | ||
| 60 | TRPM3 | 2470110 6400731 6550047 | 4667 | 0.245 | 0.4075 | No | ||
| 61 | DOCK4 | 5910102 | 4811 | 0.229 | 0.4015 | No | ||
| 62 | CHRNA2 | 5050315 | 5026 | 0.205 | 0.3915 | No | ||
| 63 | FLNC | 2360048 5050270 | 5349 | 0.171 | 0.3754 | No | ||
| 64 | MYO18B | 7100670 | 5401 | 0.167 | 0.3739 | No | ||
| 65 | BRUNOL4 | 4490452 5080451 | 5441 | 0.164 | 0.3731 | No | ||
| 66 | BRS3 | 1740403 | 5781 | 0.138 | 0.3558 | No | ||
| 67 | CYR61 | 1240408 5290026 4120452 6550008 | 5791 | 0.138 | 0.3564 | No | ||
| 68 | HOXC4 | 1940193 | 5855 | 0.134 | 0.3540 | No | ||
| 69 | B4GALT5 | 1230692 | 5916 | 0.129 | 0.3518 | No | ||
| 70 | USH1G | 2940446 | 6135 | 0.116 | 0.3408 | No | ||
| 71 | SLITRK5 | 6590292 | 6172 | 0.115 | 0.3398 | No | ||
| 72 | PPARD | 110131 | 6238 | 0.111 | 0.3371 | No | ||
| 73 | MUSK | 1190113 | 6411 | 0.101 | 0.3286 | No | ||
| 74 | OTOP2 | 610068 | 6523 | 0.096 | 0.3233 | No | ||
| 75 | RFX5 | 5420128 | 6661 | 0.091 | 0.3166 | No | ||
| 76 | VWF | 4670519 | 6858 | 0.083 | 0.3066 | No | ||
| 77 | FGF10 | 730458 840706 | 6977 | 0.079 | 0.3008 | No | ||
| 78 | NRP2 | 4070400 5860041 6650446 | 7011 | 0.077 | 0.2997 | No | ||
| 79 | RFX4 | 3710100 5340215 | 7382 | 0.065 | 0.2801 | No | ||
| 80 | A2M | 6620039 | 7534 | 0.061 | 0.2724 | No | ||
| 81 | MOBKL2C | 2640088 3830017 | 7577 | 0.060 | 0.2706 | No | ||
| 82 | TRIB1 | 2320435 | 7647 | 0.058 | 0.2673 | No | ||
| 83 | RCL1 | 1980408 2680008 3800176 | 7946 | 0.051 | 0.2515 | No | ||
| 84 | HOXA5 | 6840026 | 8304 | 0.043 | 0.2325 | No | ||
| 85 | ELP4 | 1230576 | 8484 | 0.040 | 0.2231 | No | ||
| 86 | NRG1 | 1050332 | 8525 | 0.039 | 0.2212 | No | ||
| 87 | SPATS2 | 2360066 | 8545 | 0.039 | 0.2205 | No | ||
| 88 | BDNF | 2940128 3520368 | 8647 | 0.036 | 0.2153 | No | ||
| 89 | SLC26A7 | 1850398 3830377 | 8768 | 0.033 | 0.2091 | No | ||
| 90 | FOXA1 | 3710609 | 8791 | 0.033 | 0.2081 | No | ||
| 91 | GJB1 | 1240441 | 8839 | 0.032 | 0.2058 | No | ||
| 92 | PRDM1 | 3170347 3520301 | 8950 | 0.030 | 0.2001 | No | ||
| 93 | PRRX1 | 4120193 4480390 | 9053 | 0.028 | 0.1948 | No | ||
| 94 | FGF14 | 2630390 3520075 6770048 | 9223 | 0.024 | 0.1858 | No | ||
| 95 | SRPX | 580717 1980162 | 9303 | 0.023 | 0.1817 | No | ||
| 96 | TGFBR1 | 1400148 4280020 6550711 | 9349 | 0.022 | 0.1794 | No | ||
| 97 | GRIA1 | 1340152 3780750 4920440 | 9369 | 0.022 | 0.1786 | No | ||
| 98 | LMO4 | 3800746 | 9397 | 0.021 | 0.1773 | No | ||
| 99 | SFTPC | 3290133 | 9407 | 0.021 | 0.1769 | No | ||
| 100 | CDH16 | 1450129 | 9634 | 0.017 | 0.1648 | No | ||
| 101 | PAX3 | 50551 | 9645 | 0.017 | 0.1644 | No | ||
| 102 | SPTBN4 | 4150279 5270286 | 9729 | 0.016 | 0.1600 | No | ||
| 103 | XPR1 | 1500093 | 9745 | 0.015 | 0.1593 | No | ||
| 104 | JAG1 | 3440390 | 9767 | 0.015 | 0.1583 | No | ||
| 105 | ADRA2C | 5130154 | 9912 | 0.013 | 0.1506 | No | ||
| 106 | CRLF1 | 3520092 | 10036 | 0.010 | 0.1440 | No | ||
| 107 | PDHA2 | 2630438 | 10094 | 0.010 | 0.1410 | No | ||
| 108 | HNT | 6450731 | 10146 | 0.009 | 0.1383 | No | ||
| 109 | TMPRSS5 | 4060068 | 10151 | 0.009 | 0.1381 | No | ||
| 110 | NKX2-8 | 50022 | 10156 | 0.009 | 0.1380 | No | ||
| 111 | PLAC1 | 2350594 | 10291 | 0.006 | 0.1308 | No | ||
| 112 | BET1 | 4010725 | 10335 | 0.006 | 0.1285 | No | ||
| 113 | SP7 | 1050315 | 10445 | 0.004 | 0.1226 | No | ||
| 114 | BMP5 | 4670048 | 10581 | 0.001 | 0.1153 | No | ||
| 115 | TAL1 | 7040239 | 10620 | 0.001 | 0.1132 | No | ||
| 116 | CP | 2570484 | 10734 | -0.001 | 0.1071 | No | ||
| 117 | CHRDL1 | 1580037 6380204 | 10750 | -0.001 | 0.1063 | No | ||
| 118 | CHML | 6650156 | 11003 | -0.005 | 0.0927 | No | ||
| 119 | P4HA2 | 1090546 | 11103 | -0.007 | 0.0874 | No | ||
| 120 | GTPBP1 | 2230541 4560528 | 11381 | -0.012 | 0.0724 | No | ||
| 121 | SPRY2 | 5860184 | 11448 | -0.014 | 0.0690 | No | ||
| 122 | CRYGB | 2340593 | 11547 | -0.016 | 0.0638 | No | ||
| 123 | TEAD1 | 2470551 | 11592 | -0.016 | 0.0615 | No | ||
| 124 | TGFB3 | 1070041 | 11664 | -0.017 | 0.0578 | No | ||
| 125 | ACCN1 | 2060139 2190541 | 11830 | -0.021 | 0.0490 | No | ||
| 126 | UPK2 | 60176 | 11837 | -0.021 | 0.0488 | No | ||
| 127 | EMX2 | 1660092 | 11898 | -0.022 | 0.0458 | No | ||
| 128 | MYF6 | 2690576 | 11901 | -0.022 | 0.0458 | No | ||
| 129 | HNF4G | 5720451 | 12001 | -0.025 | 0.0407 | No | ||
| 130 | HOXB9 | 2640687 | 12230 | -0.029 | 0.0285 | No | ||
| 131 | SKIL | 1090364 | 12261 | -0.030 | 0.0271 | No | ||
| 132 | HOXD3 | 6450154 | 12268 | -0.030 | 0.0270 | No | ||
| 133 | GSC | 1580685 | 12276 | -0.030 | 0.0269 | No | ||
| 134 | CHRND | 840403 2260670 | 12359 | -0.033 | 0.0227 | No | ||
| 135 | BNC1 | 5550427 | 12403 | -0.034 | 0.0206 | No | ||
| 136 | IRX3 | 4200112 | 12472 | -0.036 | 0.0172 | No | ||
| 137 | ARHGAP12 | 1050181 | 12556 | -0.038 | 0.0130 | No | ||
| 138 | SNCAIP | 4010551 | 12736 | -0.043 | 0.0036 | No | ||
| 139 | OIT3 | 1660152 | 12775 | -0.044 | 0.0019 | No | ||
| 140 | T | 6370164 | 13111 | -0.055 | -0.0158 | No | ||
| 141 | KCNIP2 | 60088 1780324 | 13132 | -0.055 | -0.0165 | No | ||
| 142 | SEMA3A | 1780064 | 13262 | -0.061 | -0.0230 | No | ||
| 143 | DHH | 2120433 | 13487 | -0.070 | -0.0346 | No | ||
| 144 | PFTK1 | 1780181 | 13767 | -0.086 | -0.0491 | No | ||
| 145 | NTRK1 | 6400538 | 13833 | -0.091 | -0.0519 | No | ||
| 146 | PTCH2 | 2900092 | 13904 | -0.096 | -0.0549 | No | ||
| 147 | ZIC3 | 380020 | 13960 | -0.100 | -0.0571 | No | ||
| 148 | ATF3 | 1940546 | 14024 | -0.105 | -0.0597 | No | ||
| 149 | KCNA2 | 5080403 | 14027 | -0.105 | -0.0590 | No | ||
| 150 | HAVCR2 | 6860592 | 14151 | -0.116 | -0.0648 | No | ||
| 151 | ITGA7 | 1230739 | 14183 | -0.120 | -0.0655 | No | ||
| 152 | AMD1 | 6290128 | 14673 | -0.188 | -0.0906 | No | ||
| 153 | LUZP1 | 940075 4280458 6180070 | 14789 | -0.213 | -0.0951 | No | ||
| 154 | RNF34 | 3800332 4590717 5670138 | 14888 | -0.232 | -0.0986 | No | ||
| 155 | EN1 | 5360168 7050025 | 15069 | -0.281 | -0.1062 | No | ||
| 156 | BCL2L2 | 2760692 6770739 | 15120 | -0.296 | -0.1066 | No | ||
| 157 | SLCO3A1 | 1050408 2370156 6110072 | 15329 | -0.360 | -0.1150 | No | ||
| 158 | SP6 | 60484 510452 2690333 | 15341 | -0.366 | -0.1128 | No | ||
| 159 | AMOTL1 | 380279 | 15349 | -0.370 | -0.1102 | No | ||
| 160 | DNTTIP1 | 5360537 | 15452 | -0.403 | -0.1126 | No | ||
| 161 | NEDD4 | 460736 1190280 | 15556 | -0.438 | -0.1147 | No | ||
| 162 | GPR85 | 5130750 5910020 | 15793 | -0.530 | -0.1234 | No | ||
| 163 | CHD2 | 4070039 | 15963 | -0.611 | -0.1278 | No | ||
| 164 | MOSPD1 | 2030170 5270156 6900142 | 16002 | -0.626 | -0.1249 | No | ||
| 165 | HHEX | 2340575 | 16159 | -0.703 | -0.1279 | No | ||
| 166 | PPP1R12A | 4070066 4590162 | 16223 | -0.748 | -0.1254 | No | ||
| 167 | PPARGC1B | 3290114 | 16381 | -0.828 | -0.1274 | No | ||
| 168 | MYADM | 4230132 | 16517 | -0.905 | -0.1276 | No | ||
| 169 | GATA3 | 6130068 | 16668 | -0.991 | -0.1280 | No | ||
| 170 | UBE2H | 1980142 2970079 | 16734 | -1.024 | -0.1235 | No | ||
| 171 | MAP2K6 | 1230056 2940204 | 16855 | -1.102 | -0.1213 | No | ||
| 172 | MBNL1 | 2640762 7100048 | 17040 | -1.263 | -0.1214 | No | ||
| 173 | BZW1 | 460270 | 17347 | -1.584 | -0.1256 | No | ||
| 174 | LDB1 | 5270601 | 17353 | -1.597 | -0.1133 | No | ||
| 175 | MCTS1 | 6130133 | 17468 | -1.732 | -0.1059 | No | ||
| 176 | SOCS2 | 4760692 | 17492 | -1.751 | -0.0934 | No | ||
| 177 | RUNX1 | 3840711 | 17573 | -1.837 | -0.0833 | No | ||
| 178 | SLIT3 | 7100132 | 17591 | -1.864 | -0.0696 | No | ||
| 179 | PPID | 2680707 | 17702 | -2.022 | -0.0597 | No | ||
| 180 | BTG1 | 4200735 6040131 6200133 | 17833 | -2.195 | -0.0495 | No | ||
| 181 | STIP1 | 6450390 | 17842 | -2.205 | -0.0327 | No | ||
| 182 | OSBPL9 | 2570373 | 17913 | -2.316 | -0.0183 | No | ||
| 183 | LUC7L | 6040156 6110411 | 18294 | -3.137 | -0.0143 | No | ||
| 184 | METAP1 | 3520632 | 18495 | -4.035 | 0.0066 | No |