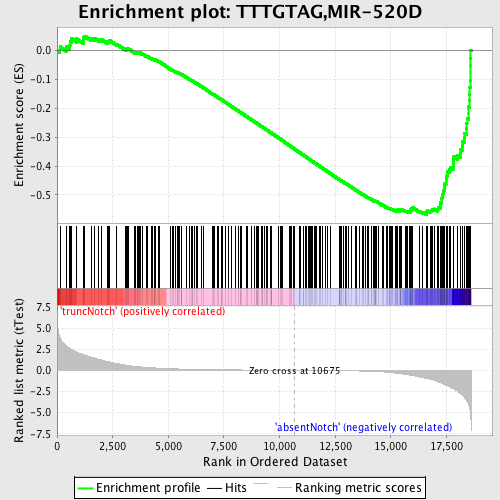
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #truncNotch_versus_absentNotch |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#truncNotch_versus_absentNotch |
| Upregulated in class | absentNotch |
| GeneSet | TTTGTAG,MIR-520D |
| Enrichment Score (ES) | -0.567601 |
| Normalized Enrichment Score (NES) | -1.7555332 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0054989704 |
| FWER p-Value | 0.043 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | HES1 | 4810280 | 129 | 3.885 | 0.0136 | No | ||
| 2 | TUSC2 | 1580253 | 401 | 2.931 | 0.0144 | No | ||
| 3 | CUEDC1 | 1660463 | 563 | 2.628 | 0.0196 | No | ||
| 4 | ETV5 | 110017 | 582 | 2.603 | 0.0324 | No | ||
| 5 | ARRDC1 | 4850095 | 639 | 2.506 | 0.0427 | No | ||
| 6 | RCE1 | 1980372 | 877 | 2.167 | 0.0413 | No | ||
| 7 | VAMP2 | 2100100 | 1175 | 1.856 | 0.0350 | No | ||
| 8 | HES5 | 4920026 | 1178 | 1.852 | 0.0447 | No | ||
| 9 | TXN2 | 2030402 | 1244 | 1.777 | 0.0506 | No | ||
| 10 | FKBP1A | 2450368 | 1524 | 1.554 | 0.0437 | No | ||
| 11 | ATXN2 | 3940017 4810242 | 1662 | 1.473 | 0.0440 | No | ||
| 12 | SON | 3120148 5860239 7040403 | 1865 | 1.303 | 0.0400 | No | ||
| 13 | U2AF1 | 5130292 | 2011 | 1.199 | 0.0384 | No | ||
| 14 | SCD | 2680441 | 2249 | 1.046 | 0.0311 | No | ||
| 15 | SEMA4G | 5890026 | 2322 | 1.010 | 0.0325 | No | ||
| 16 | UBE2E3 | 2030347 3840471 | 2353 | 0.996 | 0.0362 | No | ||
| 17 | MDM1 | 510528 1780280 2680152 2690181 3060025 3120487 4210601 4540037 4610142 5290110 6400433 6480743 | 2676 | 0.810 | 0.0230 | No | ||
| 18 | NEUROG1 | 380341 | 3084 | 0.616 | 0.0041 | No | ||
| 19 | CDC42BPB | 6350037 | 3106 | 0.605 | 0.0061 | No | ||
| 20 | EGR3 | 6940128 | 3176 | 0.582 | 0.0055 | No | ||
| 21 | HNRPA0 | 2680048 | 3219 | 0.567 | 0.0062 | No | ||
| 22 | PPP1CB | 3130411 3780270 4150180 | 3486 | 0.473 | -0.0058 | No | ||
| 23 | EIF3S10 | 1190079 3840176 | 3518 | 0.465 | -0.0050 | No | ||
| 24 | MTF2 | 50333 | 3610 | 0.438 | -0.0076 | No | ||
| 25 | CPEB2 | 4760338 | 3674 | 0.423 | -0.0088 | No | ||
| 26 | CCND1 | 460524 770309 3120576 6980398 | 3689 | 0.419 | -0.0073 | No | ||
| 27 | DCAMKL1 | 540095 2690092 | 3727 | 0.411 | -0.0072 | No | ||
| 28 | LRBA | 3450402 | 3835 | 0.388 | -0.0109 | No | ||
| 29 | GADD45A | 2900717 | 4034 | 0.346 | -0.0199 | No | ||
| 30 | SOCS5 | 3830398 7100093 | 4072 | 0.339 | -0.0201 | No | ||
| 31 | PHF6 | 3120632 3990041 | 4227 | 0.313 | -0.0268 | No | ||
| 32 | KDELR1 | 6760504 | 4291 | 0.302 | -0.0286 | No | ||
| 33 | DDIT4L | 3370292 | 4370 | 0.288 | -0.0313 | No | ||
| 34 | CDH2 | 520435 2450451 2760025 6650477 | 4395 | 0.285 | -0.0311 | No | ||
| 35 | ETS1 | 5270278 6450717 6620465 | 4410 | 0.282 | -0.0304 | No | ||
| 36 | RDX | 4200039 6550465 | 4569 | 0.258 | -0.0376 | No | ||
| 37 | SDC2 | 1980168 | 4589 | 0.255 | -0.0373 | No | ||
| 38 | NEBL | 1850369 2480286 | 5093 | 0.194 | -0.0637 | No | ||
| 39 | SCN3A | 4070309 | 5189 | 0.184 | -0.0678 | No | ||
| 40 | LRCH4 | 5220279 6510026 | 5220 | 0.181 | -0.0685 | No | ||
| 41 | RKHD3 | 6180471 | 5319 | 0.173 | -0.0729 | No | ||
| 42 | ZZZ3 | 780133 2320239 2640452 5080706 6550341 | 5399 | 0.167 | -0.0763 | No | ||
| 43 | NHLH2 | 6200021 | 5411 | 0.166 | -0.0761 | No | ||
| 44 | LRRN1 | 3290154 | 5439 | 0.164 | -0.0767 | No | ||
| 45 | ENAH | 1690292 5700300 | 5512 | 0.158 | -0.0797 | No | ||
| 46 | BTBD3 | 1980041 2510577 3190053 4200746 | 5577 | 0.153 | -0.0824 | No | ||
| 47 | GAD1 | 2360035 3140167 | 5579 | 0.153 | -0.0816 | No | ||
| 48 | RFX3 | 520121 4280519 5570519 6100184 | 5592 | 0.153 | -0.0815 | No | ||
| 49 | PIWIL4 | 3180471 | 5803 | 0.137 | -0.0922 | No | ||
| 50 | COCH | 1090136 | 5950 | 0.127 | -0.0995 | No | ||
| 51 | CASP8AP2 | 5910593 | 6044 | 0.121 | -0.1039 | No | ||
| 52 | CNTNAP2 | 360170 4280546 | 6067 | 0.120 | -0.1044 | No | ||
| 53 | ZFHX4 | 6110167 | 6106 | 0.118 | -0.1059 | No | ||
| 54 | HOXB5 | 2900538 | 6179 | 0.114 | -0.1092 | No | ||
| 55 | ATXN1 | 5550156 | 6268 | 0.109 | -0.1134 | No | ||
| 56 | GRM3 | 4540242 | 6319 | 0.106 | -0.1156 | No | ||
| 57 | HOXB6 | 4210739 | 6477 | 0.099 | -0.1236 | No | ||
| 58 | MAP3K2 | 380270 | 6589 | 0.094 | -0.1291 | No | ||
| 59 | PHLPP | 3440563 | 6987 | 0.078 | -0.1503 | No | ||
| 60 | JMJD1C | 940575 2120025 | 7022 | 0.077 | -0.1518 | No | ||
| 61 | ACTL6A | 5220543 6290541 2450093 4590332 5270722 | 7052 | 0.076 | -0.1529 | No | ||
| 62 | GPM6A | 1660044 2750152 | 7224 | 0.071 | -0.1619 | No | ||
| 63 | RFX1 | 540373 | 7232 | 0.070 | -0.1619 | No | ||
| 64 | RFX4 | 3710100 5340215 | 7382 | 0.065 | -0.1696 | No | ||
| 65 | DKK2 | 3610433 | 7393 | 0.065 | -0.1698 | No | ||
| 66 | ODC1 | 5670168 | 7414 | 0.064 | -0.1706 | No | ||
| 67 | OSBPL10 | 1980184 | 7556 | 0.060 | -0.1779 | No | ||
| 68 | SOX1 | 630026 | 7695 | 0.057 | -0.1851 | No | ||
| 69 | FBN2 | 50435 | 7842 | 0.054 | -0.1928 | No | ||
| 70 | EIF2C1 | 6900551 | 8005 | 0.050 | -0.2014 | No | ||
| 71 | GABRA1 | 130372 | 8173 | 0.046 | -0.2102 | No | ||
| 72 | KCNK1 | 1050068 | 8222 | 0.045 | -0.2126 | No | ||
| 73 | ARL4A | 6290091 | 8257 | 0.044 | -0.2142 | No | ||
| 74 | FOSL1 | 430021 | 8299 | 0.043 | -0.2162 | No | ||
| 75 | BHLHB3 | 3450438 | 8505 | 0.039 | -0.2271 | No | ||
| 76 | PTPN2 | 130315 | 8559 | 0.038 | -0.2298 | No | ||
| 77 | LPP | 1660541 | 8726 | 0.034 | -0.2387 | No | ||
| 78 | CRHBP | 5420181 | 8870 | 0.031 | -0.2463 | No | ||
| 79 | PRDM1 | 3170347 3520301 | 8950 | 0.030 | -0.2504 | No | ||
| 80 | TLOC1 | 4050309 | 8969 | 0.029 | -0.2513 | No | ||
| 81 | NRXN3 | 1400546 3360471 | 8996 | 0.029 | -0.2525 | No | ||
| 82 | SYNPR | 2810400 | 9022 | 0.028 | -0.2537 | No | ||
| 83 | HDGFRP3 | 6180451 | 9052 | 0.028 | -0.2552 | No | ||
| 84 | STX12 | 610451 | 9185 | 0.025 | -0.2622 | No | ||
| 85 | KLF11 | 6840739 | 9224 | 0.024 | -0.2641 | No | ||
| 86 | SLITRK3 | 5910041 | 9253 | 0.024 | -0.2655 | No | ||
| 87 | PTN | 5910161 | 9328 | 0.023 | -0.2694 | No | ||
| 88 | MTSS1 | 780435 2370114 | 9419 | 0.021 | -0.2742 | No | ||
| 89 | PURA | 3870156 | 9424 | 0.021 | -0.2743 | No | ||
| 90 | DACH1 | 2450593 5700592 | 9455 | 0.020 | -0.2759 | No | ||
| 91 | ABCA1 | 6290156 | 9591 | 0.018 | -0.2831 | No | ||
| 92 | CHES1 | 4570121 5700162 | 9623 | 0.017 | -0.2847 | No | ||
| 93 | SYT1 | 840364 | 9642 | 0.017 | -0.2856 | No | ||
| 94 | ABCC12 | 5550026 | 9646 | 0.017 | -0.2857 | No | ||
| 95 | GPM6B | 6220458 | 9946 | 0.012 | -0.3019 | No | ||
| 96 | CBX4 | 70332 | 10054 | 0.010 | -0.3077 | No | ||
| 97 | TBC1D15 | 870673 | 10073 | 0.010 | -0.3086 | No | ||
| 98 | NR5A2 | 1690180 6900050 | 10142 | 0.009 | -0.3122 | No | ||
| 99 | NOPE | 380156 | 10429 | 0.004 | -0.3278 | No | ||
| 100 | FBXO22 | 4760010 | 10482 | 0.003 | -0.3306 | No | ||
| 101 | SP4 | 3850176 | 10522 | 0.002 | -0.3327 | No | ||
| 102 | TAL1 | 7040239 | 10620 | 0.001 | -0.3380 | No | ||
| 103 | KIF11 | 5390139 | 10668 | 0.000 | -0.3405 | No | ||
| 104 | SRC | 580132 | 10879 | -0.003 | -0.3519 | No | ||
| 105 | TRPC4 | 6040022 | 10911 | -0.004 | -0.3536 | No | ||
| 106 | SERTAD2 | 2810673 6840450 | 10958 | -0.005 | -0.3561 | No | ||
| 107 | SEH1L | 4590563 | 11070 | -0.007 | -0.3621 | No | ||
| 108 | POLR2B | 3780138 | 11096 | -0.007 | -0.3634 | No | ||
| 109 | PPP3CA | 4760332 6760092 | 11159 | -0.008 | -0.3668 | No | ||
| 110 | NEUROG2 | 5420278 | 11188 | -0.009 | -0.3682 | No | ||
| 111 | SBF2 | 3060315 | 11288 | -0.010 | -0.3736 | No | ||
| 112 | PDZRN3 | 2340131 | 11353 | -0.012 | -0.3770 | No | ||
| 113 | GTPBP1 | 2230541 4560528 | 11381 | -0.012 | -0.3784 | No | ||
| 114 | PFN2 | 730040 5700026 | 11442 | -0.014 | -0.3816 | No | ||
| 115 | KLF12 | 1660095 4810288 5340546 6520286 | 11457 | -0.014 | -0.3823 | No | ||
| 116 | SNF1LK | 6110403 | 11552 | -0.016 | -0.3873 | No | ||
| 117 | TEAD1 | 2470551 | 11592 | -0.016 | -0.3893 | No | ||
| 118 | PDCL | 2340348 | 11671 | -0.018 | -0.3935 | No | ||
| 119 | MAFF | 4850577 5720286 | 11792 | -0.020 | -0.3999 | No | ||
| 120 | ABHD3 | 2370091 | 11845 | -0.021 | -0.4026 | No | ||
| 121 | IER5 | 2350575 | 11920 | -0.023 | -0.4065 | No | ||
| 122 | TIPARP | 50397 | 12055 | -0.026 | -0.4137 | No | ||
| 123 | ADD1 | 1500176 3060139 3390092 | 12160 | -0.028 | -0.4192 | No | ||
| 124 | HOXD3 | 6450154 | 12268 | -0.030 | -0.4249 | No | ||
| 125 | SLC35F1 | 3850164 | 12673 | -0.041 | -0.4466 | No | ||
| 126 | TMEM47 | 1660086 | 12677 | -0.041 | -0.4466 | No | ||
| 127 | KCNMA1 | 1450402 | 12701 | -0.042 | -0.4476 | No | ||
| 128 | PRPF4B | 940181 2570300 | 12711 | -0.042 | -0.4479 | No | ||
| 129 | POU4F2 | 2120195 2570022 | 12730 | -0.042 | -0.4486 | No | ||
| 130 | MEF2C | 670025 780338 | 12795 | -0.044 | -0.4519 | No | ||
| 131 | GIPC2 | 3800463 | 12796 | -0.045 | -0.4516 | No | ||
| 132 | NPTX1 | 3710736 | 12876 | -0.047 | -0.4557 | No | ||
| 133 | FMN2 | 450463 | 12958 | -0.050 | -0.4598 | No | ||
| 134 | USP33 | 780047 | 12974 | -0.050 | -0.4604 | No | ||
| 135 | NDFIP1 | 1980402 | 12977 | -0.050 | -0.4602 | No | ||
| 136 | RSBN1 | 7000487 | 12999 | -0.051 | -0.4611 | No | ||
| 137 | LIN28 | 6590672 | 13101 | -0.054 | -0.4663 | No | ||
| 138 | ADCYAP1 | 520086 | 13246 | -0.060 | -0.4738 | No | ||
| 139 | SP3 | 3840338 | 13400 | -0.066 | -0.4818 | No | ||
| 140 | NPAS4 | 2320204 4150154 | 13441 | -0.068 | -0.4836 | No | ||
| 141 | QKI | 5220093 6130044 | 13451 | -0.068 | -0.4838 | No | ||
| 142 | SNAP25 | 360520 | 13606 | -0.076 | -0.4917 | No | ||
| 143 | UBE3A | 1240152 2690438 5860609 | 13746 | -0.084 | -0.4988 | No | ||
| 144 | CSTF3 | 3850156 5570458 | 13768 | -0.086 | -0.4995 | No | ||
| 145 | DAPK1 | 5570010 | 13872 | -0.093 | -0.5046 | No | ||
| 146 | RPS6KB1 | 1450427 5080110 6200563 | 13959 | -0.099 | -0.5088 | No | ||
| 147 | BIVM | 6040546 | 13985 | -0.102 | -0.5096 | No | ||
| 148 | UBQLN2 | 430397 1170594 4070121 | 14001 | -0.103 | -0.5099 | No | ||
| 149 | HOXA7 | 5910152 | 14119 | -0.113 | -0.5157 | No | ||
| 150 | WASF2 | 4850592 | 14136 | -0.115 | -0.5159 | No | ||
| 151 | PPARGC1A | 4670040 | 14233 | -0.125 | -0.5205 | No | ||
| 152 | GNAO1 | 2510292 6590487 | 14267 | -0.129 | -0.5216 | No | ||
| 153 | PTP4A1 | 130692 | 14273 | -0.130 | -0.5212 | No | ||
| 154 | SLC1A3 | 1500070 | 14295 | -0.133 | -0.5216 | No | ||
| 155 | SERPINF2 | 2470390 2690292 | 14300 | -0.133 | -0.5211 | No | ||
| 156 | PIP5K3 | 5360112 | 14309 | -0.134 | -0.5208 | No | ||
| 157 | HTR7 | 2680176 | 14347 | -0.139 | -0.5221 | No | ||
| 158 | RNF12 | 6860717 | 14433 | -0.150 | -0.5259 | No | ||
| 159 | CLNS1A | 2970022 3190402 | 14604 | -0.175 | -0.5343 | No | ||
| 160 | USP47 | 2640497 | 14655 | -0.186 | -0.5360 | No | ||
| 161 | ZNF22 | 780546 2320131 | 14816 | -0.217 | -0.5436 | No | ||
| 162 | SLITRK2 | 2640020 | 14847 | -0.222 | -0.5440 | No | ||
| 163 | RANBP2 | 4280338 | 14930 | -0.242 | -0.5472 | No | ||
| 164 | TCF7L2 | 2190348 5220113 | 14949 | -0.248 | -0.5469 | No | ||
| 165 | UBE2L3 | 1690707 5900242 | 14994 | -0.258 | -0.5479 | No | ||
| 166 | SLC35F5 | 2360369 | 15041 | -0.272 | -0.5489 | No | ||
| 167 | SOX11 | 610279 | 15082 | -0.284 | -0.5496 | No | ||
| 168 | DNAJC6 | 4590438 5900070 | 15201 | -0.323 | -0.5543 | No | ||
| 169 | TRPS1 | 870100 | 15205 | -0.324 | -0.5528 | No | ||
| 170 | TOX | 3440053 | 15260 | -0.341 | -0.5539 | No | ||
| 171 | PDE4D | 2470528 6660014 | 15290 | -0.349 | -0.5536 | No | ||
| 172 | BRWD1 | 3130048 3140168 6400601 | 15291 | -0.349 | -0.5518 | No | ||
| 173 | CITED2 | 5670114 5130088 | 15380 | -0.379 | -0.5545 | No | ||
| 174 | NCAM1 | 3140026 | 15416 | -0.391 | -0.5544 | No | ||
| 175 | STK17B | 130091 | 15418 | -0.393 | -0.5523 | No | ||
| 176 | PRPF39 | 1090088 | 15439 | -0.398 | -0.5513 | No | ||
| 177 | ANKRD10 | 1240025 | 15441 | -0.399 | -0.5493 | No | ||
| 178 | CBX3 | 1780592 3940008 | 15485 | -0.413 | -0.5494 | No | ||
| 179 | IGF1 | 1990193 3130377 3290280 | 15657 | -0.476 | -0.5562 | No | ||
| 180 | XPO5 | 2850286 | 15709 | -0.496 | -0.5563 | No | ||
| 181 | PHF15 | 6550066 6860673 | 15765 | -0.521 | -0.5566 | No | ||
| 182 | HNRPM | 450497 4540451 | 15821 | -0.542 | -0.5567 | No | ||
| 183 | MAPKAP1 | 1660161 2450575 | 15876 | -0.566 | -0.5566 | No | ||
| 184 | SORCS1 | 60411 5890373 | 15882 | -0.570 | -0.5539 | No | ||
| 185 | MLLT10 | 1410288 3290142 | 15899 | -0.577 | -0.5517 | No | ||
| 186 | MBNL2 | 3450707 | 15937 | -0.596 | -0.5505 | No | ||
| 187 | MTA1 | 3830092 | 15943 | -0.600 | -0.5476 | No | ||
| 188 | RAI14 | 770288 1660075 | 15953 | -0.607 | -0.5449 | No | ||
| 189 | PCNP | 610768 | 15965 | -0.611 | -0.5422 | No | ||
| 190 | SFRS5 | 3450176 6350008 | 16289 | -0.778 | -0.5557 | No | ||
| 191 | SULF2 | 460086 | 16436 | -0.860 | -0.5591 | No | ||
| 192 | ACACA | 2490612 2680369 | 16594 | -0.954 | -0.5625 | Yes | ||
| 193 | SESTD1 | 1090333 3780593 | 16622 | -0.967 | -0.5589 | Yes | ||
| 194 | PDCD4 | 3360403 5690110 | 16635 | -0.974 | -0.5544 | Yes | ||
| 195 | TPST1 | 2570358 | 16763 | -1.043 | -0.5557 | Yes | ||
| 196 | MECP2 | 1940450 | 16813 | -1.075 | -0.5527 | Yes | ||
| 197 | VGLL4 | 6860463 | 16861 | -1.106 | -0.5494 | Yes | ||
| 198 | PUM1 | 6130500 | 16962 | -1.182 | -0.5485 | Yes | ||
| 199 | ATF4 | 730441 1740195 | 17104 | -1.330 | -0.5492 | Yes | ||
| 200 | CCT8 | 6220019 | 17139 | -1.366 | -0.5438 | Yes | ||
| 201 | CMTM4 | 360600 3520332 | 17217 | -1.443 | -0.5403 | Yes | ||
| 202 | PHF2 | 4060300 | 17233 | -1.466 | -0.5333 | Yes | ||
| 203 | EIF4G2 | 3800575 6860184 | 17237 | -1.470 | -0.5257 | Yes | ||
| 204 | MOCS2 | 2940497 | 17287 | -1.520 | -0.5203 | Yes | ||
| 205 | CUGBP2 | 6180121 6840139 | 17288 | -1.521 | -0.5122 | Yes | ||
| 206 | PRDM2 | 780278 1940692 | 17303 | -1.537 | -0.5048 | Yes | ||
| 207 | SMOC2 | 1170170 | 17317 | -1.553 | -0.4973 | Yes | ||
| 208 | BZW1 | 460270 | 17347 | -1.584 | -0.4904 | Yes | ||
| 209 | PURB | 5360138 | 17367 | -1.611 | -0.4829 | Yes | ||
| 210 | ADIPOR1 | 3140440 | 17414 | -1.664 | -0.4766 | Yes | ||
| 211 | SIRT1 | 1190731 | 17426 | -1.676 | -0.4683 | Yes | ||
| 212 | DHX32 | 2680138 | 17433 | -1.688 | -0.4597 | Yes | ||
| 213 | LRP11 | 1980324 | 17482 | -1.740 | -0.4530 | Yes | ||
| 214 | SMOC1 | 840039 | 17484 | -1.741 | -0.4439 | Yes | ||
| 215 | UHRF1 | 1410273 1660242 | 17494 | -1.752 | -0.4351 | Yes | ||
| 216 | FNTA | 70059 | 17541 | -1.808 | -0.4280 | Yes | ||
| 217 | ELF1 | 4780450 | 17546 | -1.813 | -0.4186 | Yes | ||
| 218 | NXF1 | 1570377 | 17625 | -1.897 | -0.4127 | Yes | ||
| 219 | ATP2A2 | 1090075 3990279 | 17677 | -1.987 | -0.4050 | Yes | ||
| 220 | MAP3K7IP2 | 2340242 | 17796 | -2.145 | -0.4000 | Yes | ||
| 221 | SCN5A | 4070017 | 17807 | -2.160 | -0.3891 | Yes | ||
| 222 | LRRC8D | 1780014 | 17820 | -2.173 | -0.3782 | Yes | ||
| 223 | ZFX | 5900400 | 17836 | -2.197 | -0.3673 | Yes | ||
| 224 | HNRPDL | 1050102 1090181 5360471 | 18013 | -2.480 | -0.3638 | Yes | ||
| 225 | NEDD9 | 1740373 4230053 | 18130 | -2.734 | -0.3556 | Yes | ||
| 226 | CUTL1 | 1570113 1770091 1850273 2100286 5340605 | 18147 | -2.771 | -0.3417 | Yes | ||
| 227 | SFRS6 | 60224 | 18213 | -2.933 | -0.3297 | Yes | ||
| 228 | NDEL1 | 2370465 | 18236 | -2.993 | -0.3150 | Yes | ||
| 229 | LUC7L | 6040156 6110411 | 18294 | -3.137 | -0.3014 | Yes | ||
| 230 | RNF44 | 5910692 | 18326 | -3.259 | -0.2858 | Yes | ||
| 231 | H3F3A | 2900086 | 18397 | -3.542 | -0.2708 | Yes | ||
| 232 | TRAM1 | 2340441 6620592 | 18409 | -3.588 | -0.2524 | Yes | ||
| 233 | ARL8B | 3710368 | 18425 | -3.654 | -0.2338 | Yes | ||
| 234 | PPP2CA | 3990113 | 18501 | -4.067 | -0.2163 | Yes | ||
| 235 | ELAVL1 | 4730497 | 18503 | -4.082 | -0.1947 | Yes | ||
| 236 | UBE3C | 3140673 | 18533 | -4.295 | -0.1735 | Yes | ||
| 237 | TMPO | 4050494 | 18542 | -4.448 | -0.1503 | Yes | ||
| 238 | CTCF | 5340017 | 18555 | -4.586 | -0.1266 | Yes | ||
| 239 | NFYB | 1850053 | 18567 | -4.754 | -0.1020 | Yes | ||
| 240 | STK4 | 2640152 | 18573 | -4.814 | -0.0767 | Yes | ||
| 241 | H3F3B | 1410300 | 18574 | -4.822 | -0.0511 | Yes | ||
| 242 | SFRS1 | 2360440 | 18577 | -4.846 | -0.0255 | Yes | ||
| 243 | PPM1D | 1690193 | 18592 | -5.188 | 0.0013 | Yes |

