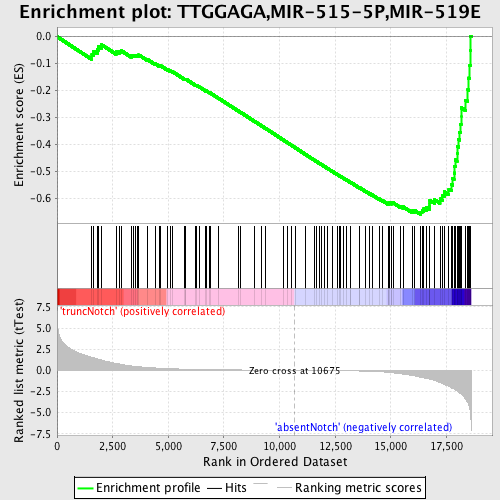
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #truncNotch_versus_absentNotch |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#truncNotch_versus_absentNotch |
| Upregulated in class | absentNotch |
| GeneSet | TTGGAGA,MIR-515-5P,MIR-519E |
| Enrichment Score (ES) | -0.6584884 |
| Normalized Enrichment Score (NES) | -1.85604 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0015973331 |
| FWER p-Value | 0.0080 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TGFBR2 | 1780711 1980537 6550398 | 1564 | 1.525 | -0.0672 | No | ||
| 2 | RIOK3 | 1980487 4480619 7040026 | 1634 | 1.489 | -0.0541 | No | ||
| 3 | SERTAD1 | 2680332 | 1805 | 1.348 | -0.0481 | No | ||
| 4 | POM121 | 70152 | 1844 | 1.319 | -0.0352 | No | ||
| 5 | EFNA3 | 6370390 | 1995 | 1.210 | -0.0297 | No | ||
| 6 | FSTL1 | 2900113 | 2650 | 0.827 | -0.0557 | No | ||
| 7 | TMEM2 | 1980037 3290020 | 2789 | 0.751 | -0.0546 | No | ||
| 8 | POU2F1 | 70577 430373 4850324 5910056 | 2892 | 0.694 | -0.0523 | No | ||
| 9 | GABBR1 | 2850519 2850735 | 3330 | 0.528 | -0.0699 | No | ||
| 10 | GDI1 | 4590400 5290358 6020687 6180446 | 3413 | 0.500 | -0.0687 | No | ||
| 11 | CSNK1G1 | 840082 1230575 3940647 | 3503 | 0.469 | -0.0682 | No | ||
| 12 | MTF2 | 50333 | 3610 | 0.438 | -0.0690 | No | ||
| 13 | CPEB2 | 4760338 | 3674 | 0.423 | -0.0676 | No | ||
| 14 | SCN8A | 380253 | 4063 | 0.341 | -0.0847 | No | ||
| 15 | ETS1 | 5270278 6450717 6620465 | 4410 | 0.282 | -0.1002 | No | ||
| 16 | UHRF2 | 60050 840280 1340746 5080091 | 4595 | 0.254 | -0.1073 | No | ||
| 17 | KCND2 | 1170128 2100112 | 4632 | 0.250 | -0.1064 | No | ||
| 18 | MLLT6 | 3870168 | 4972 | 0.210 | -0.1224 | No | ||
| 19 | ABCC3 | 4050242 | 5094 | 0.194 | -0.1267 | No | ||
| 20 | HIP2 | 2810095 3990369 4120301 | 5188 | 0.184 | -0.1296 | No | ||
| 21 | HNRPUL1 | 2120008 4920408 5570180 6370609 6770403 | 5720 | 0.143 | -0.1567 | No | ||
| 22 | SOX30 | 2690725 4730309 | 5763 | 0.140 | -0.1574 | No | ||
| 23 | CYR61 | 1240408 5290026 4120452 6550008 | 5791 | 0.138 | -0.1573 | No | ||
| 24 | MEF2D | 5690576 | 6231 | 0.112 | -0.1797 | No | ||
| 25 | SLC4A4 | 1780075 1850040 4810068 | 6282 | 0.109 | -0.1812 | No | ||
| 26 | CLSTN2 | 6900546 | 6395 | 0.102 | -0.1861 | No | ||
| 27 | NFIB | 460450 | 6690 | 0.089 | -0.2010 | No | ||
| 28 | CLCF1 | 4920176 6550750 | 6701 | 0.089 | -0.2005 | No | ||
| 29 | NUP35 | 5340408 | 6827 | 0.084 | -0.2063 | No | ||
| 30 | EPN2 | 1340451 6550239 6860020 6860403 | 6876 | 0.082 | -0.2080 | No | ||
| 31 | APLN | 6620632 | 7271 | 0.069 | -0.2285 | No | ||
| 32 | KCTD7 | 4230114 | 8140 | 0.047 | -0.2748 | No | ||
| 33 | FRAS1 | 5080170 5670347 | 8245 | 0.045 | -0.2799 | No | ||
| 34 | ACY1L2 | 3140142 | 8852 | 0.032 | -0.3123 | No | ||
| 35 | PSMD11 | 2340538 6510053 | 9169 | 0.025 | -0.3291 | No | ||
| 36 | ITSN1 | 2640577 | 9376 | 0.022 | -0.3400 | No | ||
| 37 | SLC9A2 | 4920022 5910133 | 10179 | 0.008 | -0.3832 | No | ||
| 38 | CAB39 | 5720471 | 10373 | 0.005 | -0.3936 | No | ||
| 39 | SP4 | 3850176 | 10522 | 0.002 | -0.4015 | No | ||
| 40 | ETNK2 | 4540373 | 10736 | -0.001 | -0.4130 | No | ||
| 41 | ANUBL1 | 1580113 | 11142 | -0.008 | -0.4348 | No | ||
| 42 | UBE2B | 4780497 | 11559 | -0.016 | -0.4571 | No | ||
| 43 | HOXA1 | 1190524 5420142 | 11646 | -0.017 | -0.4616 | No | ||
| 44 | MOSPD2 | 4010064 | 11786 | -0.020 | -0.4688 | No | ||
| 45 | EMX2 | 1660092 | 11898 | -0.022 | -0.4746 | No | ||
| 46 | ISL1 | 4810239 | 12040 | -0.025 | -0.4819 | No | ||
| 47 | ETV1 | 70735 2940603 5080463 | 12133 | -0.027 | -0.4866 | No | ||
| 48 | DDX3Y | 1580278 4200519 | 12371 | -0.033 | -0.4990 | No | ||
| 49 | BSN | 5270347 | 12607 | -0.039 | -0.5112 | No | ||
| 50 | TMEM47 | 1660086 | 12677 | -0.041 | -0.5145 | No | ||
| 51 | DCX | 130075 5360025 7050673 | 12733 | -0.043 | -0.5170 | No | ||
| 52 | SYNGR1 | 5130452 6650056 | 12879 | -0.047 | -0.5243 | No | ||
| 53 | RSBN1 | 7000487 | 12999 | -0.051 | -0.5301 | No | ||
| 54 | OPHN1 | 2360100 | 13176 | -0.057 | -0.5390 | No | ||
| 55 | NOTCH2 | 2570397 | 13205 | -0.058 | -0.5399 | No | ||
| 56 | SNAP25 | 360520 | 13606 | -0.076 | -0.5606 | No | ||
| 57 | ARRDC3 | 4610390 | 13611 | -0.076 | -0.5600 | No | ||
| 58 | BTG2 | 2350411 | 13859 | -0.092 | -0.5723 | No | ||
| 59 | NFATC4 | 2470735 | 14059 | -0.108 | -0.5818 | No | ||
| 60 | DGCR2 | 780041 4010731 | 14161 | -0.118 | -0.5859 | No | ||
| 61 | ATRN | 870735 3360390 | 14468 | -0.154 | -0.6007 | No | ||
| 62 | OSBPL3 | 3170164 4210196 | 14635 | -0.181 | -0.6076 | No | ||
| 63 | ABCG4 | 3440458 | 14896 | -0.235 | -0.6190 | No | ||
| 64 | PDGFRA | 2940332 | 14915 | -0.239 | -0.6173 | No | ||
| 65 | SOX12 | 1780037 | 14929 | -0.242 | -0.6153 | No | ||
| 66 | FGFR2 | 4670601 5570402 6940377 | 15016 | -0.265 | -0.6169 | No | ||
| 67 | RBL2 | 580446 1400670 | 15024 | -0.267 | -0.6143 | No | ||
| 68 | ZFP36L1 | 2510138 4120048 | 15103 | -0.292 | -0.6152 | No | ||
| 69 | TNPO1 | 730092 2260735 | 15413 | -0.390 | -0.6275 | No | ||
| 70 | DONSON | 4780541 6130010 | 15557 | -0.438 | -0.6302 | No | ||
| 71 | ATXN3 | 50156 2900095 | 15962 | -0.611 | -0.6452 | No | ||
| 72 | ZBTB1 | 5220010 | 16048 | -0.647 | -0.6424 | No | ||
| 73 | PLAGL2 | 1770148 | 16346 | -0.808 | -0.6494 | Yes | ||
| 74 | SGPL1 | 4480059 | 16404 | -0.840 | -0.6429 | Yes | ||
| 75 | RPS6KA3 | 1980707 | 16486 | -0.889 | -0.6373 | Yes | ||
| 76 | ULK2 | 50041 5050176 | 16592 | -0.953 | -0.6322 | Yes | ||
| 77 | UBE2H | 1980142 2970079 | 16734 | -1.024 | -0.6282 | Yes | ||
| 78 | SUMO2 | 70010 3610706 | 16747 | -1.033 | -0.6172 | Yes | ||
| 79 | ARHGEF3 | 3440156 | 16752 | -1.036 | -0.6057 | Yes | ||
| 80 | PUM1 | 6130500 | 16962 | -1.182 | -0.6037 | Yes | ||
| 81 | FREQ | 3140039 4060600 4560670 | 17211 | -1.438 | -0.6008 | Yes | ||
| 82 | IQGAP1 | 6660494 | 17330 | -1.572 | -0.5894 | Yes | ||
| 83 | SLC7A11 | 2850138 | 17401 | -1.655 | -0.5745 | Yes | ||
| 84 | MTHFD1L | 3360131 | 17608 | -1.882 | -0.5644 | Yes | ||
| 85 | YAF2 | 940053 | 17726 | -2.061 | -0.5474 | Yes | ||
| 86 | KPNA1 | 5270324 | 17777 | -2.121 | -0.5261 | Yes | ||
| 87 | YTHDF2 | 4060332 4610279 | 17844 | -2.212 | -0.5047 | Yes | ||
| 88 | ARID1A | 2630022 1690551 4810110 | 17864 | -2.236 | -0.4805 | Yes | ||
| 89 | CMTM6 | 6590010 | 17895 | -2.290 | -0.4562 | Yes | ||
| 90 | NCOA1 | 3610438 | 17979 | -2.418 | -0.4334 | Yes | ||
| 91 | UBAP2 | 460398 | 17988 | -2.438 | -0.4063 | Yes | ||
| 92 | ICK | 1580746 3140021 | 18048 | -2.543 | -0.3807 | Yes | ||
| 93 | EPC2 | 2470095 | 18091 | -2.651 | -0.3531 | Yes | ||
| 94 | DDX3X | 2190020 | 18122 | -2.725 | -0.3239 | Yes | ||
| 95 | RDH11 | 4810673 | 18161 | -2.803 | -0.2943 | Yes | ||
| 96 | MARCKS | 7040450 | 18164 | -2.808 | -0.2627 | Yes | ||
| 97 | RAP2C | 1690132 | 18365 | -3.418 | -0.2349 | Yes | ||
| 98 | RAP1B | 3710138 | 18463 | -3.825 | -0.1969 | Yes | ||
| 99 | MAPRE1 | 3290037 | 18494 | -4.031 | -0.1530 | Yes | ||
| 100 | RPP14 | 7040014 | 18550 | -4.485 | -0.1053 | Yes | ||
| 101 | AP3S1 | 5570044 | 18568 | -4.776 | -0.0522 | Yes | ||
| 102 | SFRS1 | 2360440 | 18577 | -4.846 | 0.0021 | Yes |

