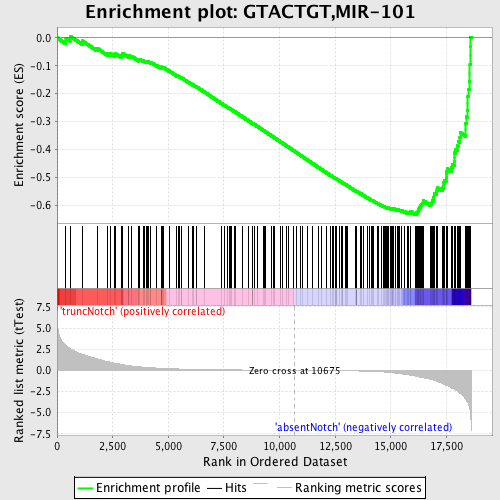
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #truncNotch_versus_absentNotch |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#truncNotch_versus_absentNotch |
| Upregulated in class | absentNotch |
| GeneSet | GTACTGT,MIR-101 |
| Enrichment Score (ES) | -0.6327581 |
| Normalized Enrichment Score (NES) | -1.9123977 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.003481572 |
| FWER p-Value | 0.0040 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SMARCD1 | 3060193 3850184 6400369 | 397 | 2.934 | -0.0016 | No | ||
| 2 | ETV5 | 110017 | 582 | 2.603 | 0.0062 | No | ||
| 3 | JUNB | 4230048 | 1138 | 1.886 | -0.0111 | No | ||
| 4 | UBE2Q1 | 610348 2350114 4210717 | 1799 | 1.355 | -0.0376 | No | ||
| 5 | BCL9 | 7100112 | 2281 | 1.030 | -0.0567 | No | ||
| 6 | BAZ2A | 730184 | 2418 | 0.958 | -0.0576 | No | ||
| 7 | RAPH1 | 6760411 | 2581 | 0.864 | -0.0605 | No | ||
| 8 | FEM1C | 60300 6130717 6980164 | 2619 | 0.844 | -0.0567 | No | ||
| 9 | FBXW7 | 4210338 7050280 | 2874 | 0.703 | -0.0657 | No | ||
| 10 | HAS2 | 5360181 | 2891 | 0.694 | -0.0619 | No | ||
| 11 | IGFBP5 | 2360592 | 2943 | 0.674 | -0.0601 | No | ||
| 12 | BZW2 | 940079 | 2954 | 0.670 | -0.0560 | No | ||
| 13 | LASS2 | 4120086 6770358 | 3203 | 0.574 | -0.0656 | No | ||
| 14 | DLG5 | 450215 | 3210 | 0.570 | -0.0620 | No | ||
| 15 | KCNH7 | 7510068 | 3349 | 0.522 | -0.0660 | No | ||
| 16 | MAGI1 | 6510500 1990239 4670471 | 3669 | 0.425 | -0.0804 | No | ||
| 17 | CPEB2 | 4760338 | 3674 | 0.423 | -0.0777 | No | ||
| 18 | CDYL | 4200142 4730397 6060400 | 3724 | 0.411 | -0.0776 | No | ||
| 19 | ING3 | 110438 2060390 | 3864 | 0.380 | -0.0825 | No | ||
| 20 | FLRT2 | 4200035 | 3942 | 0.364 | -0.0842 | No | ||
| 21 | SOX9 | 5720681 | 3998 | 0.352 | -0.0848 | No | ||
| 22 | SOCS5 | 3830398 7100093 | 4072 | 0.339 | -0.0864 | No | ||
| 23 | PREI3 | 3170066 3440075 6370427 | 4099 | 0.334 | -0.0856 | No | ||
| 24 | ACCN2 | 6450465 | 4207 | 0.317 | -0.0892 | No | ||
| 25 | NEUROD1 | 3060619 | 4212 | 0.316 | -0.0873 | No | ||
| 26 | RNF38 | 1940746 3140133 3840601 | 4472 | 0.272 | -0.0995 | No | ||
| 27 | TSC22D1 | 1340739 6040181 | 4673 | 0.244 | -0.1087 | No | ||
| 28 | PDE4A | 1190619 1340129 5720100 6370035 | 4675 | 0.243 | -0.1071 | No | ||
| 29 | MAK | 2940706 | 4678 | 0.243 | -0.1055 | No | ||
| 30 | RIPK5 | 1230647 1340114 | 4686 | 0.243 | -0.1042 | No | ||
| 31 | FOS | 1850315 | 4728 | 0.238 | -0.1048 | No | ||
| 32 | DDIT4 | 2570408 | 4765 | 0.234 | -0.1052 | No | ||
| 33 | COL10A1 | 2360307 | 5037 | 0.203 | -0.1185 | No | ||
| 34 | DCBLD2 | 5890333 | 5386 | 0.168 | -0.1363 | No | ||
| 35 | LRRN1 | 3290154 | 5439 | 0.164 | -0.1380 | No | ||
| 36 | DNMT3A | 4050068 4810717 | 5504 | 0.159 | -0.1403 | No | ||
| 37 | BTBD3 | 1980041 2510577 3190053 4200746 | 5577 | 0.153 | -0.1432 | No | ||
| 38 | MYST2 | 4540494 | 5586 | 0.153 | -0.1426 | No | ||
| 39 | DDEF1 | 1170411 4070465 | 5918 | 0.129 | -0.1597 | No | ||
| 40 | ZFHX4 | 6110167 | 6106 | 0.118 | -0.1690 | No | ||
| 41 | ATRX | 2340039 3610132 4210373 | 6109 | 0.118 | -0.1683 | No | ||
| 42 | CDH5 | 5340487 | 6263 | 0.110 | -0.1759 | No | ||
| 43 | ATXN1 | 5550156 | 6268 | 0.109 | -0.1753 | No | ||
| 44 | FZD4 | 1580520 | 6611 | 0.093 | -0.1933 | No | ||
| 45 | CPEB3 | 3940164 | 7366 | 0.066 | -0.2337 | No | ||
| 46 | TMEFF1 | 2680100 | 7521 | 0.061 | -0.2417 | No | ||
| 47 | TRIB1 | 2320435 | 7647 | 0.058 | -0.2480 | No | ||
| 48 | FLRT3 | 70685 2760497 6040519 | 7738 | 0.056 | -0.2525 | No | ||
| 49 | CDH11 | 1230133 2100180 4610110 | 7772 | 0.055 | -0.2540 | No | ||
| 50 | FBN2 | 50435 | 7842 | 0.054 | -0.2573 | No | ||
| 51 | NEGR1 | 1940731 5130170 | 7990 | 0.050 | -0.2650 | No | ||
| 52 | SIX4 | 6760022 | 8020 | 0.050 | -0.2662 | No | ||
| 53 | USP38 | 4590095 | 8328 | 0.043 | -0.2826 | No | ||
| 54 | PRKAA1 | 510156 | 8600 | 0.037 | -0.2970 | No | ||
| 55 | ROBO2 | 450136 | 8774 | 0.033 | -0.3062 | No | ||
| 56 | POGZ | 870348 7040286 | 8779 | 0.033 | -0.3062 | No | ||
| 57 | EYA1 | 1450278 5220390 | 8781 | 0.033 | -0.3060 | No | ||
| 58 | KHDRBS3 | 2120154 | 8857 | 0.032 | -0.3098 | No | ||
| 59 | TEAD3 | 2320142 | 9009 | 0.029 | -0.3178 | No | ||
| 60 | CLCN3 | 510195 780176 1660093 3830372 | 9283 | 0.023 | -0.3325 | No | ||
| 61 | ADAMTS10 | 730563 | 9309 | 0.023 | -0.3337 | No | ||
| 62 | TGFBR1 | 1400148 4280020 6550711 | 9349 | 0.022 | -0.3357 | No | ||
| 63 | THRB | 1780673 | 9619 | 0.017 | -0.3501 | No | ||
| 64 | NRK | 4590609 | 9722 | 0.016 | -0.3556 | No | ||
| 65 | FBXW11 | 6450632 | 9778 | 0.015 | -0.3584 | No | ||
| 66 | PCDH8 | 2360047 4200452 | 10056 | 0.010 | -0.3734 | No | ||
| 67 | GLRA2 | 7000019 | 10130 | 0.009 | -0.3773 | No | ||
| 68 | PACRG | 3140053 | 10301 | 0.006 | -0.3865 | No | ||
| 69 | CCNJ | 870047 | 10415 | 0.004 | -0.3926 | No | ||
| 70 | TAL1 | 7040239 | 10620 | 0.001 | -0.4036 | No | ||
| 71 | FGA | 4210056 5220239 | 10747 | -0.001 | -0.4105 | No | ||
| 72 | PUM2 | 4200441 5910446 | 10923 | -0.004 | -0.4199 | No | ||
| 73 | EXOC5 | 7000711 | 11008 | -0.006 | -0.4244 | No | ||
| 74 | PALM2 | 4120068 7100398 | 11266 | -0.010 | -0.4383 | No | ||
| 75 | RAB5A | 6520324 | 11473 | -0.014 | -0.4494 | No | ||
| 76 | CTTNBP2 | 2650167 | 11769 | -0.019 | -0.4653 | No | ||
| 77 | HNRPF | 670138 | 11877 | -0.022 | -0.4709 | No | ||
| 78 | PRKCE | 5700053 | 12129 | -0.027 | -0.4844 | No | ||
| 79 | DR1 | 5860750 | 12291 | -0.031 | -0.4929 | No | ||
| 80 | DDX3Y | 1580278 4200519 | 12371 | -0.033 | -0.4969 | No | ||
| 81 | UBE2D1 | 1850400 | 12406 | -0.034 | -0.4986 | No | ||
| 82 | FZD6 | 360288 | 12516 | -0.037 | -0.5042 | No | ||
| 83 | LRRC4 | 3850041 | 12533 | -0.037 | -0.5048 | No | ||
| 84 | PLXNB1 | 6220273 | 12548 | -0.038 | -0.5053 | No | ||
| 85 | ASPN | 2450021 | 12576 | -0.038 | -0.5065 | No | ||
| 86 | PRPF4B | 940181 2570300 | 12711 | -0.042 | -0.5135 | No | ||
| 87 | HOXD4 | 5900301 | 12792 | -0.044 | -0.5176 | No | ||
| 88 | MPPE1 | 5080364 7050739 | 12843 | -0.046 | -0.5200 | No | ||
| 89 | NUDT11 | 3840239 | 12846 | -0.046 | -0.5198 | No | ||
| 90 | NDFIP1 | 1980402 | 12977 | -0.050 | -0.5265 | No | ||
| 91 | PTGS2 | 2510301 3170369 | 12987 | -0.051 | -0.5266 | No | ||
| 92 | PPFIA4 | 630446 | 13062 | -0.053 | -0.5303 | No | ||
| 93 | MYRIP | 1580471 | 13410 | -0.066 | -0.5486 | No | ||
| 94 | SMARCA1 | 4230315 | 13439 | -0.067 | -0.5497 | No | ||
| 95 | QKI | 5220093 6130044 | 13451 | -0.068 | -0.5498 | No | ||
| 96 | LRRN3 | 1450133 | 13465 | -0.069 | -0.5501 | No | ||
| 97 | RGS1 | 4060347 4540181 | 13477 | -0.069 | -0.5502 | No | ||
| 98 | MMP15 | 5900717 | 13652 | -0.079 | -0.5591 | No | ||
| 99 | CAV3 | 1770519 | 13659 | -0.079 | -0.5589 | No | ||
| 100 | ZCCHC2 | 460592 | 13750 | -0.085 | -0.5632 | No | ||
| 101 | PLCG1 | 6020369 | 13938 | -0.098 | -0.5727 | No | ||
| 102 | MARK1 | 450484 | 14055 | -0.108 | -0.5782 | No | ||
| 103 | KIF1A | 450632 | 14148 | -0.116 | -0.5824 | No | ||
| 104 | ABCC5 | 2100600 5050692 | 14184 | -0.120 | -0.5835 | No | ||
| 105 | ADCY6 | 450364 6290670 6940286 | 14219 | -0.123 | -0.5845 | No | ||
| 106 | DUSP1 | 6860121 | 14399 | -0.144 | -0.5932 | No | ||
| 107 | PURG | 1190593 | 14447 | -0.151 | -0.5948 | No | ||
| 108 | SLC39A10 | 4150347 | 14577 | -0.171 | -0.6006 | No | ||
| 109 | USP47 | 2640497 | 14655 | -0.186 | -0.6035 | No | ||
| 110 | ERBB2IP | 580253 1090672 | 14707 | -0.196 | -0.6049 | No | ||
| 111 | SLC1A1 | 520372 6420059 | 14777 | -0.210 | -0.6072 | No | ||
| 112 | RASD2 | 3360010 | 14823 | -0.218 | -0.6082 | No | ||
| 113 | CDK6 | 4920253 | 14861 | -0.226 | -0.6087 | No | ||
| 114 | RIN2 | 2450184 | 14893 | -0.234 | -0.6088 | No | ||
| 115 | STAG2 | 4540132 | 14978 | -0.254 | -0.6116 | No | ||
| 116 | CEBPA | 5690519 | 15048 | -0.275 | -0.6135 | No | ||
| 117 | SOX11 | 610279 | 15082 | -0.284 | -0.6133 | No | ||
| 118 | SPRED1 | 6940706 | 15086 | -0.286 | -0.6115 | No | ||
| 119 | NLK | 2030010 2450041 | 15117 | -0.295 | -0.6111 | No | ||
| 120 | BICD2 | 1090411 | 15223 | -0.330 | -0.6146 | No | ||
| 121 | PDE4D | 2470528 6660014 | 15290 | -0.349 | -0.6158 | No | ||
| 122 | LRRFIP2 | 670575 730152 6590438 | 15327 | -0.359 | -0.6153 | No | ||
| 123 | PAPOLG | 2470400 | 15405 | -0.388 | -0.6168 | No | ||
| 124 | AEBP2 | 2970603 | 15482 | -0.413 | -0.6182 | No | ||
| 125 | DAG1 | 460053 610341 | 15592 | -0.453 | -0.6210 | No | ||
| 126 | EZH2 | 6130605 6380524 | 15735 | -0.506 | -0.6252 | No | ||
| 127 | GPR85 | 5130750 5910020 | 15793 | -0.530 | -0.6247 | No | ||
| 128 | ANKRD17 | 2120402 3850035 6400044 | 15875 | -0.565 | -0.6253 | No | ||
| 129 | STC1 | 360161 | 15888 | -0.571 | -0.6220 | No | ||
| 130 | PANK3 | 3060687 | 16087 | -0.660 | -0.6283 | Yes | ||
| 131 | LMNB1 | 5890292 6020008 | 16146 | -0.695 | -0.6267 | Yes | ||
| 132 | DOT1L | 3850068 | 16217 | -0.744 | -0.6254 | Yes | ||
| 133 | SLC30A7 | 2230463 | 16218 | -0.745 | -0.6203 | Yes | ||
| 134 | PHLDA1 | 2450020 | 16230 | -0.750 | -0.6158 | Yes | ||
| 135 | DYRK1A | 3190181 | 16263 | -0.763 | -0.6124 | Yes | ||
| 136 | SFRS5 | 3450176 6350008 | 16289 | -0.778 | -0.6084 | Yes | ||
| 137 | ZFAND3 | 2760504 | 16316 | -0.794 | -0.6044 | Yes | ||
| 138 | TXNDC4 | 450537 | 16348 | -0.809 | -0.6006 | Yes | ||
| 139 | CDK5R1 | 3870161 | 16378 | -0.826 | -0.5966 | Yes | ||
| 140 | SGPL1 | 4480059 | 16404 | -0.840 | -0.5922 | Yes | ||
| 141 | PIP5K1C | 5890164 6510148 | 16455 | -0.872 | -0.5890 | Yes | ||
| 142 | CACNB2 | 1500095 7330707 | 16463 | -0.877 | -0.5834 | Yes | ||
| 143 | REV3L | 1090717 | 16793 | -1.065 | -0.5940 | Yes | ||
| 144 | TERF2 | 3840044 | 16848 | -1.093 | -0.5895 | Yes | ||
| 145 | UBE2A | 4010563 | 16854 | -1.102 | -0.5822 | Yes | ||
| 146 | GJA1 | 5220731 | 16916 | -1.150 | -0.5777 | Yes | ||
| 147 | APP | 2510053 | 16927 | -1.158 | -0.5704 | Yes | ||
| 148 | NACA | 520326 5890673 | 16940 | -1.167 | -0.5631 | Yes | ||
| 149 | JDP2 | 2360500 | 16969 | -1.189 | -0.5565 | Yes | ||
| 150 | MBNL1 | 2640762 7100048 | 17040 | -1.263 | -0.5517 | Yes | ||
| 151 | OGT | 2360131 4610333 | 17074 | -1.298 | -0.5446 | Yes | ||
| 152 | RBBP7 | 430113 450450 2370309 | 17109 | -1.334 | -0.5374 | Yes | ||
| 153 | BCL2L11 | 780044 4200601 | 17311 | -1.546 | -0.5378 | Yes | ||
| 154 | UBE2D3 | 3190452 | 17364 | -1.609 | -0.5296 | Yes | ||
| 155 | PURB | 5360138 | 17367 | -1.611 | -0.5188 | Yes | ||
| 156 | ZNF24 | 3830750 | 17425 | -1.674 | -0.5104 | Yes | ||
| 157 | RXRB | 1780040 5340438 | 17485 | -1.741 | -0.5018 | Yes | ||
| 158 | SOCS2 | 4760692 | 17492 | -1.751 | -0.4902 | Yes | ||
| 159 | TULP4 | 2320364 | 17496 | -1.755 | -0.4784 | Yes | ||
| 160 | CBL | 6380068 | 17537 | -1.799 | -0.4683 | Yes | ||
| 161 | TLK2 | 1170161 | 17708 | -2.029 | -0.4637 | Yes | ||
| 162 | SLC19A2 | 6200750 | 17765 | -2.108 | -0.4524 | Yes | ||
| 163 | UBE2D2 | 4590671 6510446 | 17855 | -2.229 | -0.4421 | Yes | ||
| 164 | ARID1A | 2630022 1690551 4810110 | 17864 | -2.236 | -0.4273 | Yes | ||
| 165 | GNB1 | 2120397 | 17866 | -2.237 | -0.4121 | Yes | ||
| 166 | RRM1 | 4150433 | 17904 | -2.297 | -0.3984 | Yes | ||
| 167 | MTMR2 | 2260148 3990139 | 18000 | -2.460 | -0.3868 | Yes | ||
| 168 | ICK | 1580746 3140021 | 18048 | -2.543 | -0.3721 | Yes | ||
| 169 | PPFIA1 | 430176 3610524 | 18105 | -2.694 | -0.3568 | Yes | ||
| 170 | DDX3X | 2190020 | 18122 | -2.725 | -0.3391 | Yes | ||
| 171 | RAB1A | 2370671 | 18364 | -3.405 | -0.3290 | Yes | ||
| 172 | RAP2C | 1690132 | 18365 | -3.418 | -0.3057 | Yes | ||
| 173 | EMP1 | 4120438 | 18396 | -3.541 | -0.2832 | Yes | ||
| 174 | RANBP9 | 4670685 | 18429 | -3.680 | -0.2599 | Yes | ||
| 175 | RAP1B | 3710138 | 18463 | -3.825 | -0.2356 | Yes | ||
| 176 | KPNB1 | 1690138 | 18464 | -3.826 | -0.2096 | Yes | ||
| 177 | EED | 2320373 | 18506 | -4.095 | -0.1839 | Yes | ||
| 178 | SRPK2 | 6380341 | 18529 | -4.282 | -0.1559 | Yes | ||
| 179 | CTCF | 5340017 | 18555 | -4.586 | -0.1260 | Yes | ||
| 180 | SPOP | 450035 | 18556 | -4.589 | -0.0948 | Yes | ||
| 181 | AP3S1 | 5570044 | 18568 | -4.776 | -0.0629 | Yes | ||
| 182 | WSB1 | 870563 | 18569 | -4.783 | -0.0303 | Yes | ||
| 183 | STK4 | 2640152 | 18573 | -4.814 | 0.0023 | Yes |

