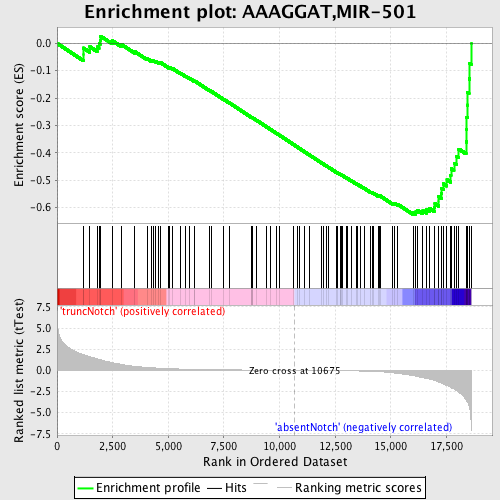
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #truncNotch_versus_absentNotch |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#truncNotch_versus_absentNotch |
| Upregulated in class | absentNotch |
| GeneSet | AAAGGAT,MIR-501 |
| Enrichment Score (ES) | -0.6254032 |
| Normalized Enrichment Score (NES) | -1.7603599 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0053781546 |
| FWER p-Value | 0.039 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | HES5 | 4920026 | 1178 | 1.852 | -0.0402 | No | ||
| 2 | TNNI2 | 5910739 | 1180 | 1.850 | -0.0169 | No | ||
| 3 | DNAJB12 | 2030148 | 1472 | 1.592 | -0.0125 | No | ||
| 4 | UBE2Q1 | 610348 2350114 4210717 | 1799 | 1.355 | -0.0130 | No | ||
| 5 | SLC25A3 | 670112 | 1884 | 1.291 | -0.0012 | No | ||
| 6 | H2AFX | 3520082 | 1932 | 1.252 | 0.0120 | No | ||
| 7 | SFRS2 | 50707 380593 | 1955 | 1.237 | 0.0265 | No | ||
| 8 | TAF5L | 5690152 | 2489 | 0.914 | 0.0092 | No | ||
| 9 | HAS2 | 5360181 | 2891 | 0.694 | -0.0037 | No | ||
| 10 | PPP1CB | 3130411 3780270 4150180 | 3486 | 0.473 | -0.0298 | No | ||
| 11 | KIF1C | 2480484 | 4060 | 0.341 | -0.0564 | No | ||
| 12 | PHF6 | 3120632 3990041 | 4227 | 0.313 | -0.0614 | No | ||
| 13 | CLK2 | 510079 2630736 | 4345 | 0.293 | -0.0640 | No | ||
| 14 | NFIL3 | 4070377 | 4428 | 0.278 | -0.0649 | No | ||
| 15 | RDX | 4200039 6550465 | 4569 | 0.258 | -0.0692 | No | ||
| 16 | KCND2 | 1170128 2100112 | 4632 | 0.250 | -0.0694 | No | ||
| 17 | HOXB8 | 3710450 | 5022 | 0.205 | -0.0878 | No | ||
| 18 | CTDSP1 | 6650215 6940731 | 5055 | 0.200 | -0.0870 | No | ||
| 19 | SCN3A | 4070309 | 5189 | 0.184 | -0.0919 | No | ||
| 20 | CLK1 | 1780551 2100102 2340347 2480347 | 5560 | 0.155 | -0.1099 | No | ||
| 21 | GIF | 2260056 | 5786 | 0.138 | -0.1203 | No | ||
| 22 | KCNRG | 3460315 3940072 | 5963 | 0.126 | -0.1282 | No | ||
| 23 | SLITRK5 | 6590292 | 6172 | 0.115 | -0.1380 | No | ||
| 24 | CUTL2 | 2940692 6220687 | 6189 | 0.114 | -0.1374 | No | ||
| 25 | ALS2 | 3130546 6400070 | 6862 | 0.083 | -0.1727 | No | ||
| 26 | RGL1 | 1500097 6900450 | 6958 | 0.079 | -0.1768 | No | ||
| 27 | PITX2 | 870537 2690139 | 7472 | 0.063 | -0.2037 | No | ||
| 28 | CART1 | 770338 | 7751 | 0.056 | -0.2180 | No | ||
| 29 | NFASC | 1940619 2470156 4590168 6840524 | 8737 | 0.034 | -0.2708 | No | ||
| 30 | WDFY3 | 3610041 4560333 4570273 | 8747 | 0.034 | -0.2708 | No | ||
| 31 | LRRC1 | 2450097 6180746 | 8760 | 0.034 | -0.2710 | No | ||
| 32 | ROBO2 | 450136 | 8774 | 0.033 | -0.2713 | No | ||
| 33 | CHODL | 7050039 | 8982 | 0.029 | -0.2821 | No | ||
| 34 | PURA | 3870156 | 9424 | 0.021 | -0.3057 | No | ||
| 35 | RET | 3850746 | 9609 | 0.018 | -0.3154 | No | ||
| 36 | SATB2 | 2470181 | 9856 | 0.014 | -0.3285 | No | ||
| 37 | RCN1 | 2480041 | 10005 | 0.011 | -0.3363 | No | ||
| 38 | NR2F2 | 3170609 3310577 | 10615 | 0.001 | -0.3692 | No | ||
| 39 | PNN | 6650504 | 10785 | -0.002 | -0.3783 | No | ||
| 40 | SEPHS1 | 4230725 | 10890 | -0.004 | -0.3839 | No | ||
| 41 | NPR3 | 1580239 | 11102 | -0.007 | -0.3952 | No | ||
| 42 | ZIC4 | 1500082 | 11366 | -0.012 | -0.4092 | No | ||
| 43 | PIK3AP1 | 6760008 | 11867 | -0.022 | -0.4360 | No | ||
| 44 | SYT7 | 2450528 3390500 3610687 | 11973 | -0.024 | -0.4413 | No | ||
| 45 | PRKCE | 5700053 | 12129 | -0.027 | -0.4494 | No | ||
| 46 | GRB10 | 6980082 | 12216 | -0.029 | -0.4536 | No | ||
| 47 | PLXNB1 | 6220273 | 12548 | -0.038 | -0.4710 | No | ||
| 48 | MYB | 1660494 5860451 6130706 | 12609 | -0.039 | -0.4738 | No | ||
| 49 | ATP6V1H | 7050717 | 12719 | -0.042 | -0.4791 | No | ||
| 50 | DCX | 130075 5360025 7050673 | 12733 | -0.043 | -0.4793 | No | ||
| 51 | MEF2C | 670025 780338 | 12795 | -0.044 | -0.4820 | No | ||
| 52 | CELSR2 | 2320148 6220152 | 12822 | -0.045 | -0.4828 | No | ||
| 53 | RSBN1 | 7000487 | 12999 | -0.051 | -0.4917 | No | ||
| 54 | LPIN1 | 1770358 3120059 | 13043 | -0.052 | -0.4934 | No | ||
| 55 | ADCYAP1 | 520086 | 13246 | -0.060 | -0.5035 | No | ||
| 56 | QKI | 5220093 6130044 | 13451 | -0.068 | -0.5137 | No | ||
| 57 | SLC35B3 | 3780433 5080725 | 13517 | -0.072 | -0.5163 | No | ||
| 58 | JUN | 840170 | 13623 | -0.077 | -0.5210 | No | ||
| 59 | SYNC1 | 580114 | 13821 | -0.090 | -0.5305 | No | ||
| 60 | NR4A3 | 2900021 5860095 5910039 | 14100 | -0.112 | -0.5441 | No | ||
| 61 | MYLK | 4010600 7000364 | 14162 | -0.118 | -0.5459 | No | ||
| 62 | SENP3 | 4200082 | 14236 | -0.125 | -0.5482 | No | ||
| 63 | BCLAF1 | 2030239 5720278 | 14438 | -0.151 | -0.5572 | No | ||
| 64 | UBAP1 | 4280059 | 14512 | -0.160 | -0.5591 | No | ||
| 65 | MAP3K8 | 2940286 | 14514 | -0.161 | -0.5571 | No | ||
| 66 | SOX11 | 610279 | 15082 | -0.284 | -0.5841 | No | ||
| 67 | USP12 | 6130735 6620280 | 15158 | -0.309 | -0.5843 | No | ||
| 68 | ELAVL4 | 50735 3360086 5220167 | 15306 | -0.352 | -0.5878 | No | ||
| 69 | PHF16 | 50373 7200161 | 16004 | -0.627 | -0.6175 | Yes | ||
| 70 | MAP2K1 | 840739 | 16129 | -0.683 | -0.6156 | Yes | ||
| 71 | ACADSB | 520170 | 16207 | -0.733 | -0.6105 | Yes | ||
| 72 | RAB22A | 110500 3830707 | 16443 | -0.865 | -0.6122 | Yes | ||
| 73 | ACACA | 2490612 2680369 | 16594 | -0.954 | -0.6083 | Yes | ||
| 74 | SRR | 1190465 3800494 | 16737 | -1.027 | -0.6030 | Yes | ||
| 75 | TRIM39 | 6550441 | 16949 | -1.173 | -0.5996 | Yes | ||
| 76 | TOMM70A | 780114 | 16982 | -1.212 | -0.5860 | Yes | ||
| 77 | VDAC2 | 7000215 | 17152 | -1.378 | -0.5777 | Yes | ||
| 78 | ZC3H7A | 1690215 | 17153 | -1.378 | -0.5603 | Yes | ||
| 79 | NFIX | 2450152 | 17270 | -1.505 | -0.5476 | Yes | ||
| 80 | CUGBP2 | 6180121 6840139 | 17288 | -1.521 | -0.5293 | Yes | ||
| 81 | CUL1 | 1990632 | 17375 | -1.620 | -0.5135 | Yes | ||
| 82 | AMMECR1 | 3800435 | 17524 | -1.786 | -0.4989 | Yes | ||
| 83 | SOX4 | 2260091 | 17681 | -1.992 | -0.4822 | Yes | ||
| 84 | SNAP29 | 6380239 | 17706 | -2.025 | -0.4579 | Yes | ||
| 85 | PPP2R5E | 70671 2120056 | 17840 | -2.202 | -0.4373 | Yes | ||
| 86 | SGPP1 | 3930707 | 17964 | -2.395 | -0.4137 | Yes | ||
| 87 | ADIPOR2 | 1510035 | 18055 | -2.558 | -0.3862 | Yes | ||
| 88 | BCL6 | 940100 | 18384 | -3.485 | -0.3599 | Yes | ||
| 89 | PDK1 | 5290427 | 18402 | -3.564 | -0.3158 | Yes | ||
| 90 | LEPROTL1 | 6900110 | 18410 | -3.591 | -0.2709 | Yes | ||
| 91 | RNF11 | 3990068 5080458 | 18437 | -3.723 | -0.2253 | Yes | ||
| 92 | PHC1 | 520195 | 18445 | -3.750 | -0.1783 | Yes | ||
| 93 | RABGEF1 | 4010497 | 18516 | -4.165 | -0.1295 | Yes | ||
| 94 | CD164 | 3830594 | 18548 | -4.479 | -0.0746 | Yes | ||
| 95 | UBE4B | 780008 3610154 | 18612 | -6.192 | 0.0002 | Yes |

