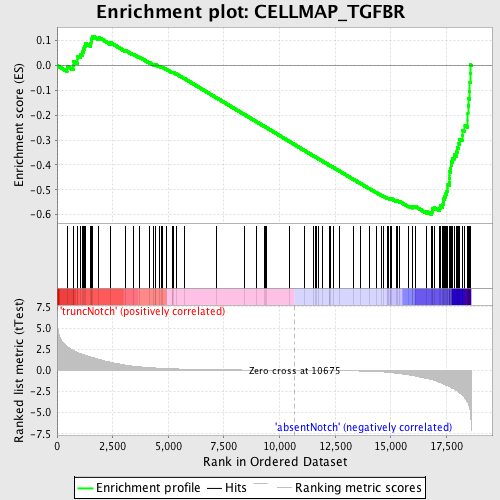
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #truncNotch_versus_absentNotch |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#truncNotch_versus_absentNotch |
| Upregulated in class | absentNotch |
| GeneSet | CELLMAP_TGFBR |
| Enrichment Score (ES) | -0.59917736 |
| Normalized Enrichment Score (NES) | -1.7017827 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.2510869 |
| FWER p-Value | 0.397 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ANAPC7 | 5260274 | 446 | 2.815 | -0.0029 | No | ||
| 2 | ARRB2 | 2060441 | 722 | 2.368 | -0.0000 | No | ||
| 3 | DAXX | 4010647 | 729 | 2.363 | 0.0174 | No | ||
| 4 | HSPA8 | 2120315 1050132 6550746 | 930 | 2.090 | 0.0223 | No | ||
| 5 | PIK3R2 | 1660193 | 932 | 2.087 | 0.0379 | No | ||
| 6 | CDK4 | 540075 4540600 | 1065 | 1.953 | 0.0455 | No | ||
| 7 | JUNB | 4230048 | 1138 | 1.886 | 0.0557 | No | ||
| 8 | TP53 | 6130707 | 1179 | 1.852 | 0.0675 | No | ||
| 9 | ANAPC2 | 6660438 | 1234 | 1.790 | 0.0780 | No | ||
| 10 | SMAD3 | 6450671 | 1267 | 1.764 | 0.0895 | No | ||
| 11 | CAMK2D | 2370685 | 1511 | 1.563 | 0.0882 | No | ||
| 12 | FKBP1A | 2450368 | 1524 | 1.554 | 0.0992 | No | ||
| 13 | TGFBR2 | 1780711 1980537 6550398 | 1564 | 1.525 | 0.1085 | No | ||
| 14 | STK11IP | 6980735 | 1600 | 1.506 | 0.1180 | No | ||
| 15 | FZR1 | 6220504 6660670 | 1881 | 1.294 | 0.1125 | No | ||
| 16 | BTRC | 3170131 | 2385 | 0.978 | 0.0927 | No | ||
| 17 | CAMK2G | 630097 | 3064 | 0.623 | 0.0608 | No | ||
| 18 | TGFB1 | 1940162 | 3412 | 0.500 | 0.0458 | No | ||
| 19 | CCND1 | 460524 770309 3120576 6980398 | 3689 | 0.419 | 0.0340 | No | ||
| 20 | SNIP1 | 5360092 | 4165 | 0.323 | 0.0108 | No | ||
| 21 | CD44 | 3990072 4850671 5860411 6860148 7050551 | 4341 | 0.293 | 0.0035 | No | ||
| 22 | ETS1 | 5270278 6450717 6620465 | 4410 | 0.282 | 0.0020 | No | ||
| 23 | AXIN1 | 3360358 6940451 | 4441 | 0.277 | 0.0024 | No | ||
| 24 | SDC2 | 1980168 | 4589 | 0.255 | -0.0036 | No | ||
| 25 | RUNX2 | 1400193 4230215 | 4693 | 0.241 | -0.0073 | No | ||
| 26 | FOS | 1850315 | 4728 | 0.238 | -0.0074 | No | ||
| 27 | SNX1 | 3190670 | 4911 | 0.217 | -0.0156 | No | ||
| 28 | FOXO3 | 2510484 4480451 | 5175 | 0.186 | -0.0284 | No | ||
| 29 | ENG | 4280270 | 5196 | 0.183 | -0.0281 | No | ||
| 30 | ZEB2 | 4120070 4150215 | 5232 | 0.180 | -0.0287 | No | ||
| 31 | SNX6 | 6200086 | 5357 | 0.170 | -0.0341 | No | ||
| 32 | AR | 6380167 | 5708 | 0.144 | -0.0519 | No | ||
| 33 | ANAPC10 | 870086 1170037 2260129 | 7177 | 0.072 | -0.1307 | No | ||
| 34 | CAV1 | 870025 | 8401 | 0.041 | -0.1964 | No | ||
| 35 | CAMK2B | 2760041 | 8961 | 0.030 | -0.2264 | No | ||
| 36 | SMAD7 | 430377 | 9300 | 0.023 | -0.2445 | No | ||
| 37 | TGFBR1 | 1400148 4280020 6550711 | 9349 | 0.022 | -0.2469 | No | ||
| 38 | NFYC | 1400746 | 9433 | 0.021 | -0.2513 | No | ||
| 39 | NUP153 | 7000452 | 10430 | 0.004 | -0.3050 | No | ||
| 40 | XPO1 | 540707 | 11124 | -0.008 | -0.3424 | No | ||
| 41 | HOXA9 | 610494 4730040 5130601 | 11544 | -0.016 | -0.3649 | No | ||
| 42 | CCNB2 | 6510528 | 11625 | -0.017 | -0.3691 | No | ||
| 43 | TGFB3 | 1070041 | 11664 | -0.017 | -0.3711 | No | ||
| 44 | SMAD6 | 870504 | 11766 | -0.019 | -0.3764 | No | ||
| 45 | CITED1 | 2350670 | 11934 | -0.023 | -0.3852 | No | ||
| 46 | FOSB | 1940142 | 12252 | -0.030 | -0.4021 | No | ||
| 47 | SKIL | 1090364 | 12261 | -0.030 | -0.4023 | No | ||
| 48 | PRKAR1B | 6130411 | 12273 | -0.030 | -0.4027 | No | ||
| 49 | UBE2D1 | 1850400 | 12406 | -0.034 | -0.4096 | No | ||
| 50 | TGFBR3 | 5290577 | 12693 | -0.042 | -0.4247 | No | ||
| 51 | ESR1 | 4060372 5860193 | 13327 | -0.063 | -0.4584 | No | ||
| 52 | JUN | 840170 | 13623 | -0.077 | -0.4738 | No | ||
| 53 | ATF3 | 1940546 | 14024 | -0.105 | -0.4946 | No | ||
| 54 | YAP1 | 4230577 | 14357 | -0.140 | -0.5115 | No | ||
| 55 | MAP3K7IP1 | 2120095 5890452 6040167 | 14572 | -0.170 | -0.5218 | No | ||
| 56 | CDC23 | 3190593 | 14662 | -0.186 | -0.5252 | No | ||
| 57 | CDK6 | 4920253 | 14861 | -0.226 | -0.5342 | No | ||
| 58 | CDKN1A | 4050088 6400706 | 14886 | -0.231 | -0.5337 | No | ||
| 59 | ZFYVE9 | 6980288 7100017 | 14979 | -0.254 | -0.5368 | No | ||
| 60 | UBE2L3 | 1690707 5900242 | 14994 | -0.258 | -0.5356 | No | ||
| 61 | RBL2 | 580446 1400670 | 15024 | -0.267 | -0.5352 | No | ||
| 62 | MYC | 380541 4670170 | 15244 | -0.337 | -0.5445 | No | ||
| 63 | VDR | 130156 510438 | 15287 | -0.348 | -0.5441 | No | ||
| 64 | ATF2 | 2260348 2480441 | 15390 | -0.383 | -0.5468 | No | ||
| 65 | MEF2A | 2350041 3360128 6040180 | 15805 | -0.535 | -0.5651 | No | ||
| 66 | SNW1 | 4010736 | 15957 | -0.608 | -0.5687 | No | ||
| 67 | SMURF2 | 1940161 | 15992 | -0.622 | -0.5659 | No | ||
| 68 | PRKAR2A | 2340136 | 16096 | -0.664 | -0.5665 | No | ||
| 69 | RBX1 | 2120010 2340047 | 16618 | -0.965 | -0.5873 | No | ||
| 70 | CDK2 | 130484 2260301 4010088 5050110 | 16838 | -1.088 | -0.5910 | Yes | ||
| 71 | MAP2K6 | 1230056 2940204 | 16855 | -1.102 | -0.5836 | Yes | ||
| 72 | EID2 | 3130176 | 16882 | -1.128 | -0.5765 | Yes | ||
| 73 | CCNE1 | 110064 | 16946 | -1.172 | -0.5711 | Yes | ||
| 74 | COPS5 | 1340722 5050215 | 17180 | -1.406 | -0.5731 | Yes | ||
| 75 | DAB2 | 60309 | 17215 | -1.442 | -0.5642 | Yes | ||
| 76 | CDC16 | 1940706 | 17321 | -1.556 | -0.5581 | Yes | ||
| 77 | UBE2D3 | 3190452 | 17364 | -1.609 | -0.5483 | Yes | ||
| 78 | CUL1 | 1990632 | 17375 | -1.620 | -0.5367 | Yes | ||
| 79 | CDC25A | 3800184 | 17420 | -1.670 | -0.5265 | Yes | ||
| 80 | MAPK14 | 5290731 | 17476 | -1.736 | -0.5165 | Yes | ||
| 81 | SNX4 | 1980358 | 17507 | -1.769 | -0.5048 | Yes | ||
| 82 | HDAC1 | 2850670 | 17539 | -1.803 | -0.4929 | Yes | ||
| 83 | FNTA | 70059 | 17541 | -1.808 | -0.4794 | Yes | ||
| 84 | TFDP1 | 1980112 | 17630 | -1.906 | -0.4698 | Yes | ||
| 85 | SP1 | 6590017 | 17641 | -1.918 | -0.4560 | Yes | ||
| 86 | PRKCD | 770592 | 17650 | -1.931 | -0.4419 | Yes | ||
| 87 | SNX2 | 2470068 3360364 6510064 | 17652 | -1.934 | -0.4274 | Yes | ||
| 88 | ANAPC5 | 730164 7100685 | 17694 | -2.010 | -0.4145 | Yes | ||
| 89 | SMAD2 | 4200592 | 17704 | -2.023 | -0.3998 | Yes | ||
| 90 | UBE2E1 | 4230064 1090440 | 17734 | -2.074 | -0.3858 | Yes | ||
| 91 | NFYA | 5860368 | 17785 | -2.132 | -0.3725 | Yes | ||
| 92 | UBE2D2 | 4590671 6510446 | 17855 | -2.229 | -0.3595 | Yes | ||
| 93 | MAP3K7 | 6040068 | 17946 | -2.361 | -0.3466 | Yes | ||
| 94 | NCOA1 | 3610438 | 17979 | -2.418 | -0.3302 | Yes | ||
| 95 | MAPK8 | 2640195 | 18042 | -2.526 | -0.3145 | Yes | ||
| 96 | CREBBP | 5690035 7040050 | 18081 | -2.625 | -0.2969 | Yes | ||
| 97 | RB1 | 5900338 | 18205 | -2.912 | -0.2816 | Yes | ||
| 98 | ZEB1 | 6840162 | 18215 | -2.938 | -0.2601 | Yes | ||
| 99 | PIAS1 | 5340301 | 18332 | -3.281 | -0.2417 | Yes | ||
| 100 | KPNB1 | 1690138 | 18464 | -3.826 | -0.2200 | Yes | ||
| 101 | RBL1 | 3130372 | 18465 | -3.829 | -0.1913 | Yes | ||
| 102 | ANAPC4 | 4540338 | 18469 | -3.854 | -0.1625 | Yes | ||
| 103 | STRAP | 1340170 | 18490 | -4.009 | -0.1334 | Yes | ||
| 104 | SMAD4 | 5670519 | 18544 | -4.465 | -0.1028 | Yes | ||
| 105 | CTCF | 5340017 | 18555 | -4.586 | -0.0689 | Yes | ||
| 106 | NFYB | 1850053 | 18567 | -4.754 | -0.0337 | Yes | ||
| 107 | ANAPC1 | 6350162 | 18575 | -4.838 | 0.0022 | Yes |