Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch_repos |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#absentNotch_versus_wtNotch_repos |
| Upregulated in class | absentNotch |
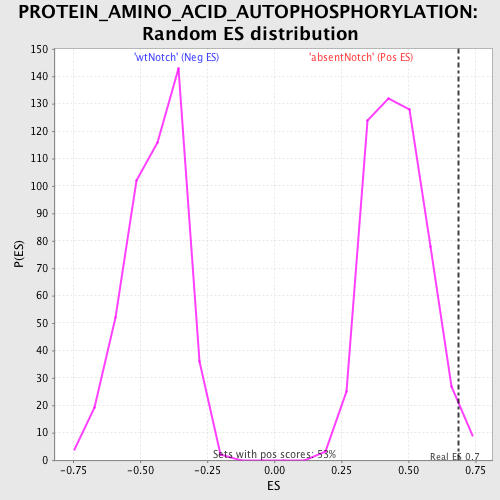
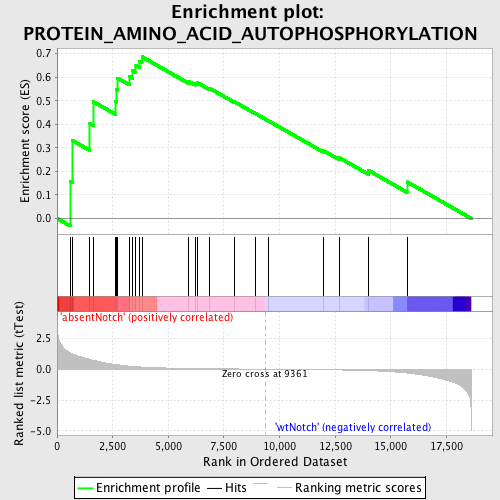
| GeneSet | PROTEIN_AMINO_ACID_AUTOPHOSPHORYLATION |
| Enrichment Score (ES) | 0.686418 |
| Normalized Enrichment Score (NES) | 1.5020826 |
| Nominal p-value | 0.017110266 |
| FDR q-value | 0.79081917 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
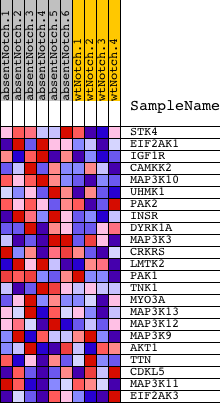
| 1 | STK4 | 2640152 | 591 | 1.319 | 0.1577 | Yes | ||
| 2 | EIF2AK1 | 2470301 | 680 | 1.246 | 0.3318 | Yes | ||
| 3 | IGF1R | 3360494 | 1474 | 0.786 | 0.4020 | Yes | ||
| 4 | CAMKK2 | 5270047 | 1634 | 0.711 | 0.4956 | Yes | ||
| 5 | MAP3K10 | 3360168 4850180 | 2618 | 0.381 | 0.4975 | Yes | ||
| 6 | UHMK1 | 3850670 6450064 | 2675 | 0.364 | 0.5468 | Yes | ||
| 7 | PAK2 | 360438 7050068 | 2712 | 0.358 | 0.5963 | Yes | ||
| 8 | INSR | 1190504 | 3250 | 0.247 | 0.6028 | Yes | ||
| 9 | DYRK1A | 3190181 | 3402 | 0.226 | 0.6271 | Yes | ||
| 10 | MAP3K3 | 610685 | 3541 | 0.206 | 0.6493 | Yes | ||
| 11 | CRKRS | 510537 2510551 | 3713 | 0.187 | 0.6670 | Yes | ||
| 12 | LMTK2 | 6650692 | 3816 | 0.174 | 0.6864 | Yes | ||
| 13 | PAK1 | 4540315 | 5902 | 0.061 | 0.5831 | No | ||
| 14 | TNK1 | 4730279 | 6207 | 0.053 | 0.5744 | No | ||
| 15 | MYO3A | 5290494 | 6299 | 0.051 | 0.5769 | No | ||
| 16 | MAP3K13 | 3190017 | 6831 | 0.040 | 0.5540 | No | ||
| 17 | MAP3K12 | 2470373 6420162 | 7980 | 0.020 | 0.4951 | No | ||
| 18 | MAP3K9 | 3290707 | 8924 | 0.006 | 0.4452 | No | ||
| 19 | AKT1 | 5290746 | 9501 | -0.002 | 0.4145 | No | ||
| 20 | TTN | 2320161 4670056 6550026 | 11975 | -0.042 | 0.2876 | No | ||
| 21 | CDKL5 | 2450070 | 12704 | -0.060 | 0.2571 | No | ||
| 22 | MAP3K11 | 7000039 | 13987 | -0.115 | 0.2047 | No | ||
| 23 | EIF2AK3 | 4280161 | 15732 | -0.308 | 0.1551 | No |