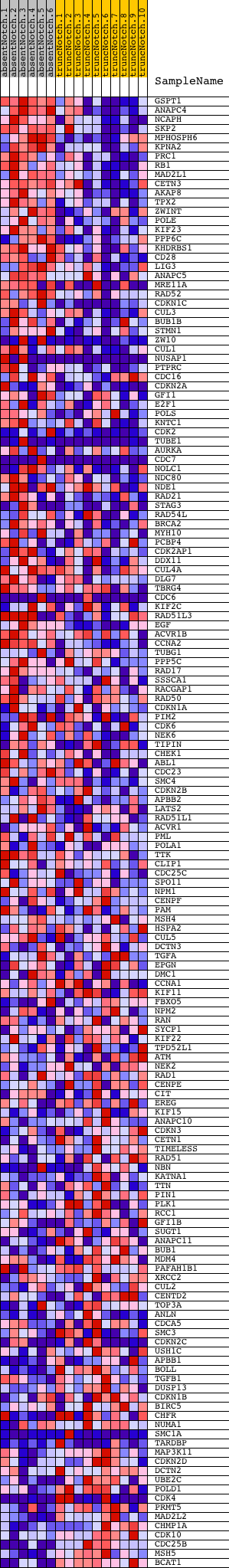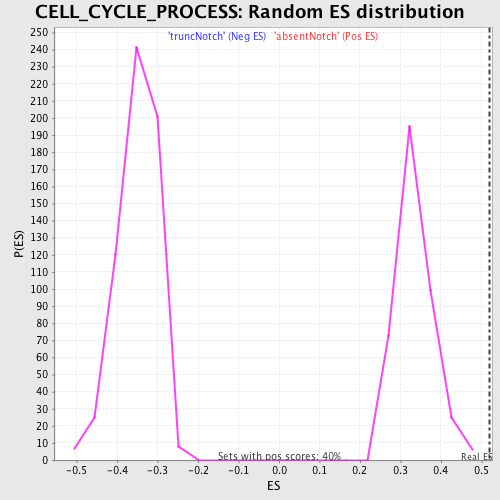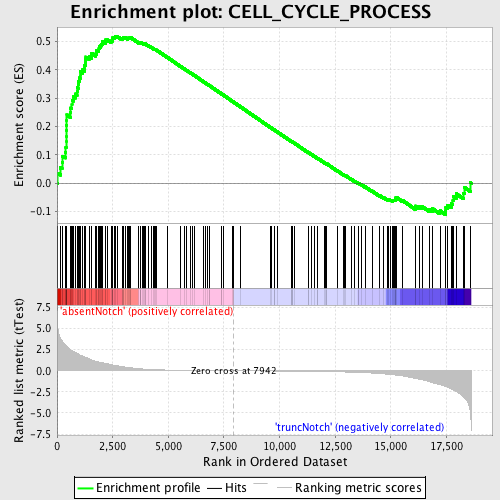
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch_repos |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#absentNotch_versus_truncNotch_repos |
| Upregulated in class | absentNotch |
| GeneSet | CELL_CYCLE_PROCESS |
| Enrichment Score (ES) | 0.51885146 |
| Normalized Enrichment Score (NES) | 1.5538954 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.61589146 |
| FWER p-Value | 0.999 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | GSPT1 | 5420050 | 18 | 5.469 | 0.0356 | Yes | ||
| 2 | ANAPC4 | 4540338 | 147 | 3.854 | 0.0544 | Yes | ||
| 3 | NCAPH | 6220435 | 224 | 3.506 | 0.0737 | Yes | ||
| 4 | SKP2 | 360711 380093 4810368 | 246 | 3.435 | 0.0956 | Yes | ||
| 5 | MPHOSPH6 | 5570164 | 370 | 3.011 | 0.1090 | Yes | ||
| 6 | KPNA2 | 3450114 | 394 | 2.961 | 0.1276 | Yes | ||
| 7 | PRC1 | 870092 5890204 | 406 | 2.929 | 0.1466 | Yes | ||
| 8 | RB1 | 5900338 | 411 | 2.912 | 0.1658 | Yes | ||
| 9 | MAD2L1 | 4480725 | 417 | 2.886 | 0.1849 | Yes | ||
| 10 | CETN3 | 3170402 | 424 | 2.872 | 0.2037 | Yes | ||
| 11 | AKAP8 | 4210026 | 436 | 2.839 | 0.2221 | Yes | ||
| 12 | TPX2 | 6420324 | 442 | 2.830 | 0.2408 | Yes | ||
| 13 | ZWINT | 6940670 | 591 | 2.496 | 0.2494 | Yes | ||
| 14 | POLE | 6020538 | 605 | 2.478 | 0.2653 | Yes | ||
| 15 | KIF23 | 5570112 | 668 | 2.363 | 0.2777 | Yes | ||
| 16 | PPP6C | 2450670 | 671 | 2.361 | 0.2934 | Yes | ||
| 17 | KHDRBS1 | 1240403 6040040 | 741 | 2.247 | 0.3047 | Yes | ||
| 18 | CD28 | 1400739 4210093 | 821 | 2.145 | 0.3148 | Yes | ||
| 19 | LIG3 | 2940300 | 896 | 2.055 | 0.3245 | Yes | ||
| 20 | ANAPC5 | 730164 7100685 | 922 | 2.010 | 0.3366 | Yes | ||
| 21 | MRE11A | 3850601 4670332 | 947 | 1.971 | 0.3485 | Yes | ||
| 22 | RAD52 | 110093 | 970 | 1.925 | 0.3601 | Yes | ||
| 23 | CDKN1C | 6520577 | 996 | 1.891 | 0.3714 | Yes | ||
| 24 | CUL3 | 1850520 | 1037 | 1.848 | 0.3816 | Yes | ||
| 25 | BUB1B | 1450288 | 1041 | 1.842 | 0.3938 | Yes | ||
| 26 | STMN1 | 1990717 | 1146 | 1.733 | 0.3997 | Yes | ||
| 27 | ZW10 | 2900735 3520687 | 1231 | 1.639 | 0.4061 | Yes | ||
| 28 | CUL1 | 1990632 | 1241 | 1.620 | 0.4165 | Yes | ||
| 29 | NUSAP1 | 940048 3120435 | 1271 | 1.583 | 0.4255 | Yes | ||
| 30 | PTPRC | 130402 5290148 | 1282 | 1.574 | 0.4355 | Yes | ||
| 31 | CDC16 | 1940706 | 1295 | 1.556 | 0.4452 | Yes | ||
| 32 | CDKN2A | 4670215 4760047 | 1438 | 1.404 | 0.4469 | Yes | ||
| 33 | GFI1 | 730180 | 1550 | 1.293 | 0.4495 | Yes | ||
| 34 | E2F1 | 5360093 | 1556 | 1.284 | 0.4578 | Yes | ||
| 35 | POLS | 5890438 | 1733 | 1.130 | 0.4559 | Yes | ||
| 36 | KNTC1 | 430079 | 1764 | 1.098 | 0.4616 | Yes | ||
| 37 | CDK2 | 130484 2260301 4010088 5050110 | 1778 | 1.088 | 0.4682 | Yes | ||
| 38 | TUBE1 | 2360079 6520685 | 1877 | 1.028 | 0.4697 | Yes | ||
| 39 | AURKA | 780537 | 1880 | 1.026 | 0.4765 | Yes | ||
| 40 | CDC7 | 4060546 4850041 | 1905 | 1.007 | 0.4819 | Yes | ||
| 41 | NOLC1 | 2350195 | 1964 | 0.983 | 0.4853 | Yes | ||
| 42 | NDC80 | 4120465 | 1990 | 0.969 | 0.4905 | Yes | ||
| 43 | NDE1 | 1580242 3830097 | 2028 | 0.950 | 0.4948 | Yes | ||
| 44 | RAD21 | 1990278 | 2041 | 0.945 | 0.5005 | Yes | ||
| 45 | STAG3 | 3830112 | 2167 | 0.869 | 0.4995 | Yes | ||
| 46 | RAD54L | 2970041 | 2168 | 0.869 | 0.5053 | Yes | ||
| 47 | BRCA2 | 4280372 | 2264 | 0.810 | 0.5056 | Yes | ||
| 48 | MYH10 | 2640673 | 2426 | 0.718 | 0.5017 | Yes | ||
| 49 | PCBP4 | 6350722 | 2474 | 0.693 | 0.5037 | Yes | ||
| 50 | CDK2AP1 | 2340156 | 2497 | 0.677 | 0.5071 | Yes | ||
| 51 | DDX11 | 6590671 | 2506 | 0.672 | 0.5111 | Yes | ||
| 52 | CUL4A | 1170088 2320008 2470278 | 2507 | 0.671 | 0.5156 | Yes | ||
| 53 | DLG7 | 3120041 | 2590 | 0.639 | 0.5155 | Yes | ||
| 54 | TBRG4 | 3610239 6370594 | 2606 | 0.629 | 0.5189 | Yes | ||
| 55 | CDC6 | 4570296 5360600 | 2713 | 0.578 | 0.5170 | No | ||
| 56 | KIF2C | 6940082 | 2928 | 0.487 | 0.5086 | No | ||
| 57 | RAD51L3 | 2230609 2810286 3190008 3390053 4230168 | 2935 | 0.484 | 0.5115 | No | ||
| 58 | EGF | 5220154 | 2963 | 0.474 | 0.5133 | No | ||
| 59 | ACVR1B | 3610446 5570195 | 2980 | 0.471 | 0.5155 | No | ||
| 60 | CCNA2 | 5290075 | 3056 | 0.440 | 0.5144 | No | ||
| 61 | TUBG1 | 3140204 | 3184 | 0.397 | 0.5102 | No | ||
| 62 | PPP5C | 3130047 | 3194 | 0.394 | 0.5123 | No | ||
| 63 | RAD17 | 5220739 | 3245 | 0.377 | 0.5121 | No | ||
| 64 | SSSCA1 | 1240402 780309 | 3284 | 0.361 | 0.5125 | No | ||
| 65 | RACGAP1 | 3990162 6620736 | 3299 | 0.354 | 0.5141 | No | ||
| 66 | RAD50 | 4050184 6770746 | 3675 | 0.246 | 0.4954 | No | ||
| 67 | CDKN1A | 4050088 6400706 | 3730 | 0.231 | 0.4941 | No | ||
| 68 | PIM2 | 840136 5720692 7100577 | 3732 | 0.231 | 0.4956 | No | ||
| 69 | CDK6 | 4920253 | 3755 | 0.226 | 0.4959 | No | ||
| 70 | NEK6 | 3360687 | 3818 | 0.213 | 0.4939 | No | ||
| 71 | TIPIN | 1570301 2480113 | 3903 | 0.197 | 0.4907 | No | ||
| 72 | CHEK1 | 50167 1940300 | 3941 | 0.188 | 0.4900 | No | ||
| 73 | ABL1 | 1050593 2030050 4010114 | 3953 | 0.186 | 0.4906 | No | ||
| 74 | CDC23 | 3190593 | 3954 | 0.186 | 0.4919 | No | ||
| 75 | SMC4 | 5910240 | 4118 | 0.158 | 0.4841 | No | ||
| 76 | CDKN2B | 6020040 | 4239 | 0.141 | 0.4785 | No | ||
| 77 | APBB2 | 5130403 | 4352 | 0.129 | 0.4733 | No | ||
| 78 | LATS2 | 4480593 6020494 | 4384 | 0.125 | 0.4725 | No | ||
| 79 | RAD51L1 | 4010112 5860435 | 4409 | 0.122 | 0.4720 | No | ||
| 80 | ACVR1 | 6840671 | 4475 | 0.115 | 0.4692 | No | ||
| 81 | PML | 50093 2190435 2450402 3840082 | 4978 | 0.078 | 0.4426 | No | ||
| 82 | POLA1 | 4760541 | 5556 | 0.053 | 0.4117 | No | ||
| 83 | TTK | 3800129 | 5743 | 0.047 | 0.4019 | No | ||
| 84 | CLIP1 | 2850162 6770397 | 5800 | 0.045 | 0.3992 | No | ||
| 85 | CDC25C | 2570673 4760161 6520707 | 6003 | 0.040 | 0.3885 | No | ||
| 86 | SPO11 | 540288 4780239 | 6065 | 0.038 | 0.3854 | No | ||
| 87 | NPM1 | 4730427 | 6182 | 0.034 | 0.3794 | No | ||
| 88 | CENPF | 5050088 | 6564 | 0.026 | 0.3589 | No | ||
| 89 | PAM | 5290528 | 6683 | 0.023 | 0.3527 | No | ||
| 90 | MSH4 | 3940068 | 6757 | 0.021 | 0.3488 | No | ||
| 91 | HSPA2 | 6420592 | 6832 | 0.020 | 0.3450 | No | ||
| 92 | CUL5 | 450142 | 6866 | 0.019 | 0.3433 | No | ||
| 93 | DCTN3 | 6840131 | 7367 | 0.010 | 0.3163 | No | ||
| 94 | TGFA | 1570332 | 7388 | 0.009 | 0.3153 | No | ||
| 95 | EPGN | 6980450 | 7462 | 0.008 | 0.3114 | No | ||
| 96 | DMC1 | 450341 | 7875 | 0.001 | 0.2890 | No | ||
| 97 | CCNA1 | 6290113 | 7918 | 0.000 | 0.2868 | No | ||
| 98 | KIF11 | 5390139 | 7948 | -0.000 | 0.2852 | No | ||
| 99 | FBXO5 | 2630551 | 8235 | -0.005 | 0.2697 | No | ||
| 100 | NPM2 | 6040139 | 9601 | -0.028 | 0.1960 | No | ||
| 101 | RAN | 2260446 4590647 | 9637 | -0.029 | 0.1943 | No | ||
| 102 | SYCP1 | 2570010 | 9755 | -0.032 | 0.1881 | No | ||
| 103 | KIF22 | 1190368 | 9789 | -0.032 | 0.1866 | No | ||
| 104 | TPD52L1 | 4590711 5570575 | 9902 | -0.035 | 0.1807 | No | ||
| 105 | ATM | 3610110 4050524 | 10515 | -0.048 | 0.1479 | No | ||
| 106 | NEK2 | 2120520 | 10547 | -0.049 | 0.1465 | No | ||
| 107 | RAD1 | 4200551 | 10569 | -0.049 | 0.1457 | No | ||
| 108 | CENPE | 2850022 | 10578 | -0.049 | 0.1456 | No | ||
| 109 | CIT | 2370601 | 10658 | -0.051 | 0.1417 | No | ||
| 110 | EREG | 50519 4920129 | 11287 | -0.067 | 0.1081 | No | ||
| 111 | KIF15 | 610731 4070731 6520014 | 11317 | -0.068 | 0.1070 | No | ||
| 112 | ANAPC10 | 870086 1170037 2260129 | 11439 | -0.072 | 0.1009 | No | ||
| 113 | CDKN3 | 110520 | 11558 | -0.076 | 0.0950 | No | ||
| 114 | CETN1 | 6020056 | 11690 | -0.081 | 0.0885 | No | ||
| 115 | TIMELESS | 3710315 | 11724 | -0.082 | 0.0872 | No | ||
| 116 | RAD51 | 6110450 6980280 | 12021 | -0.093 | 0.0718 | No | ||
| 117 | NBN | 730538 2470619 4780594 | 12073 | -0.095 | 0.0697 | No | ||
| 118 | KATNA1 | 6290048 | 12122 | -0.098 | 0.0678 | No | ||
| 119 | TTN | 2320161 4670056 6550026 | 12124 | -0.098 | 0.0684 | No | ||
| 120 | PIN1 | 2970408 | 12605 | -0.123 | 0.0432 | No | ||
| 121 | PLK1 | 1780369 2640121 | 12890 | -0.143 | 0.0287 | No | ||
| 122 | RCC1 | 2480358 | 12929 | -0.146 | 0.0277 | No | ||
| 123 | GFI1B | 4570131 | 12974 | -0.149 | 0.0263 | No | ||
| 124 | SUGT1 | 1690670 | 13218 | -0.167 | 0.0142 | No | ||
| 125 | ANAPC11 | 1780601 | 13373 | -0.179 | 0.0071 | No | ||
| 126 | BUB1 | 5390270 | 13543 | -0.197 | -0.0008 | No | ||
| 127 | MDM4 | 4070504 4780008 | 13562 | -0.200 | -0.0004 | No | ||
| 128 | PAFAH1B1 | 4230333 6420121 6450066 | 13670 | -0.213 | -0.0048 | No | ||
| 129 | XRCC2 | 4920706 | 13839 | -0.232 | -0.0123 | No | ||
| 130 | CUL2 | 4200278 | 14153 | -0.274 | -0.0274 | No | ||
| 131 | CENTD2 | 60408 2510156 6100494 | 14505 | -0.332 | -0.0442 | No | ||
| 132 | TOP3A | 7000435 | 14683 | -0.366 | -0.0514 | No | ||
| 133 | ANLN | 1780113 2470537 6840047 | 14839 | -0.399 | -0.0571 | No | ||
| 134 | CDCA5 | 5670131 | 14899 | -0.414 | -0.0575 | No | ||
| 135 | SMC3 | 870546 1400411 5700039 6450577 | 14995 | -0.436 | -0.0598 | No | ||
| 136 | CDKN2C | 5050750 5130148 | 15096 | -0.464 | -0.0621 | No | ||
| 137 | USH1C | 1570372 4230364 | 15107 | -0.468 | -0.0595 | No | ||
| 138 | APBB1 | 2690338 | 15164 | -0.486 | -0.0593 | No | ||
| 139 | BOLL | 6100743 | 15200 | -0.500 | -0.0578 | No | ||
| 140 | TGFB1 | 1940162 | 15204 | -0.500 | -0.0547 | No | ||
| 141 | DUSP13 | 6400152 | 15219 | -0.508 | -0.0520 | No | ||
| 142 | CDKN1B | 3800025 6450044 | 15233 | -0.512 | -0.0493 | No | ||
| 143 | BIRC5 | 110408 580014 1770632 | 15518 | -0.608 | -0.0606 | No | ||
| 144 | CHFR | 3390050 6450300 6450736 | 16102 | -0.901 | -0.0862 | No | ||
| 145 | NUMA1 | 6520576 | 16107 | -0.904 | -0.0804 | No | ||
| 146 | SMC1A | 3060600 5700148 5890113 6370154 | 16276 | -1.001 | -0.0828 | No | ||
| 147 | TARDBP | 870390 2350093 | 16405 | -1.070 | -0.0826 | No | ||
| 148 | MAP3K11 | 7000039 | 16729 | -1.287 | -0.0915 | No | ||
| 149 | CDKN2D | 6040035 | 16870 | -1.413 | -0.0896 | No | ||
| 150 | DCTN2 | 540471 3780717 | 17212 | -1.640 | -0.0971 | No | ||
| 151 | UBE2C | 6130017 | 17457 | -1.873 | -0.0978 | No | ||
| 152 | POLD1 | 4830026 | 17474 | -1.881 | -0.0861 | No | ||
| 153 | CDK4 | 540075 4540600 | 17551 | -1.953 | -0.0772 | No | ||
| 154 | PRMT5 | 2760180 4590072 | 17722 | -2.145 | -0.0720 | No | ||
| 155 | MAD2L2 | 1240358 | 17792 | -2.237 | -0.0608 | No | ||
| 156 | CHMP1A | 5550441 | 17809 | -2.253 | -0.0466 | No | ||
| 157 | CDK10 | 60164 1190717 3130128 3990044 | 17940 | -2.453 | -0.0373 | No | ||
| 158 | CDC25B | 6940102 | 18270 | -3.071 | -0.0346 | No | ||
| 159 | MSH5 | 1340019 4730333 | 18322 | -3.221 | -0.0158 | No | ||
| 160 | BCAT1 | 3290128 4050408 | 18570 | -4.735 | 0.0025 | No |