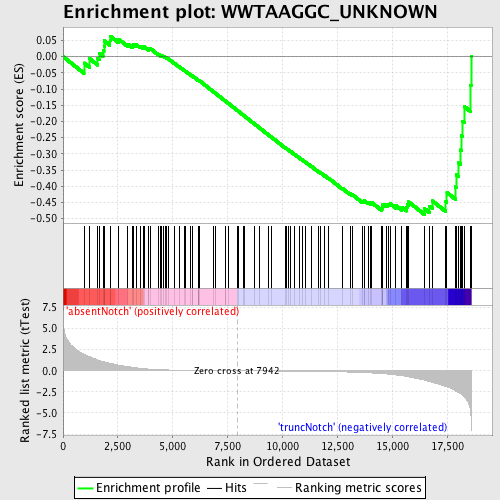
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch_repos |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#absentNotch_versus_truncNotch_repos |
| Upregulated in class | truncNotch |
| GeneSet | WWTAAGGC_UNKNOWN |
| Enrichment Score (ES) | -0.487265 |
| Normalized Enrichment Score (NES) | -1.3130968 |
| Nominal p-value | 0.043624163 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RBMS1 | 6400014 | 954 | 1.950 | -0.0196 | No | ||
| 2 | CYCS | 2030072 | 1212 | 1.657 | -0.0064 | No | ||
| 3 | MBNL1 | 2640762 7100048 | 1576 | 1.263 | -0.0054 | No | ||
| 4 | JDP2 | 2360500 | 1647 | 1.189 | 0.0103 | No | ||
| 5 | PI4K2B | 2480215 4610563 | 1821 | 1.065 | 0.0183 | No | ||
| 6 | SLC16A6 | 1690156 | 1865 | 1.035 | 0.0329 | No | ||
| 7 | BRD4 | 580471 610255 730537 1090132 1410204 1990242 2680088 3290019 4280706 | 1874 | 1.030 | 0.0494 | No | ||
| 8 | BBX | 6520112 | 2137 | 0.886 | 0.0497 | No | ||
| 9 | BHLHB2 | 7040603 | 2152 | 0.878 | 0.0633 | No | ||
| 10 | CS | 5080600 | 2538 | 0.656 | 0.0532 | No | ||
| 11 | SH2D1A | 4760025 | 2949 | 0.479 | 0.0389 | No | ||
| 12 | ANKRD10 | 1240025 | 3175 | 0.399 | 0.0333 | No | ||
| 13 | NCAM1 | 3140026 | 3200 | 0.391 | 0.0384 | No | ||
| 14 | PDE4D | 2470528 6660014 | 3326 | 0.349 | 0.0373 | No | ||
| 15 | SPAG7 | 4560524 | 3517 | 0.290 | 0.0318 | No | ||
| 16 | SLC41A1 | 1230528 | 3655 | 0.251 | 0.0285 | No | ||
| 17 | SOX12 | 1780037 | 3687 | 0.242 | 0.0308 | No | ||
| 18 | JMJD2A | 3610128 4200373 | 3905 | 0.197 | 0.0223 | No | ||
| 19 | FGFRL1 | 6370047 | 3907 | 0.197 | 0.0255 | No | ||
| 20 | HDAC9 | 1990010 2260133 | 3964 | 0.185 | 0.0255 | No | ||
| 21 | NRAS | 6900577 | 4328 | 0.132 | 0.0080 | No | ||
| 22 | MYLK | 4010600 7000364 | 4454 | 0.118 | 0.0032 | No | ||
| 23 | SHOX2 | 3190438 6450059 | 4481 | 0.115 | 0.0037 | No | ||
| 24 | NFATC4 | 2470735 | 4557 | 0.108 | 0.0014 | No | ||
| 25 | CPNE1 | 3440131 | 4655 | 0.100 | -0.0022 | No | ||
| 26 | ERG | 50154 1770739 | 4698 | 0.096 | -0.0029 | No | ||
| 27 | FOXQ1 | 4850088 | 4796 | 0.090 | -0.0067 | No | ||
| 28 | SFXN4 | 60750 4560059 | 5087 | 0.072 | -0.0212 | No | ||
| 29 | RORB | 6400035 | 5290 | 0.063 | -0.0310 | No | ||
| 30 | HOXD8 | 3710156 | 5509 | 0.055 | -0.0419 | No | ||
| 31 | SIX1 | 6760471 | 5576 | 0.052 | -0.0446 | No | ||
| 32 | SLC2A4 | 540441 | 5797 | 0.045 | -0.0558 | No | ||
| 33 | DCX | 130075 5360025 7050673 | 5883 | 0.043 | -0.0597 | No | ||
| 34 | TNP1 | 3120048 | 5894 | 0.042 | -0.0595 | No | ||
| 35 | ATP12A | 2450056 | 6161 | 0.035 | -0.0733 | No | ||
| 36 | BNC2 | 4810603 | 6167 | 0.035 | -0.0730 | No | ||
| 37 | DLL1 | 1770377 | 6175 | 0.035 | -0.0728 | No | ||
| 38 | C1QTNF3 | 3290138 6020278 | 6195 | 0.034 | -0.0733 | No | ||
| 39 | MYL9 | 4210750 7050138 | 6857 | 0.019 | -0.1087 | No | ||
| 40 | SLC16A1 | 50433 | 6944 | 0.018 | -0.1130 | No | ||
| 41 | MEIS1 | 1400575 | 7380 | 0.010 | -0.1364 | No | ||
| 42 | CFL2 | 380239 6200368 | 7413 | 0.009 | -0.1379 | No | ||
| 43 | SLCO1C1 | 5390494 | 7531 | 0.007 | -0.1441 | No | ||
| 44 | FGF17 | 3130022 | 7964 | -0.000 | -0.1675 | No | ||
| 45 | NR2F2 | 3170609 3310577 | 8001 | -0.001 | -0.1694 | No | ||
| 46 | OTX1 | 2510215 | 8204 | -0.004 | -0.1802 | No | ||
| 47 | LHX1 | 1690100 | 8265 | -0.005 | -0.1834 | No | ||
| 48 | GPR22 | 1850035 | 8703 | -0.013 | -0.2068 | No | ||
| 49 | PAX3 | 50551 | 8971 | -0.017 | -0.2209 | No | ||
| 50 | NKX2-2 | 4150731 | 9365 | -0.024 | -0.2418 | No | ||
| 51 | PHKA1 | 2810162 4120040 6040722 | 9501 | -0.027 | -0.2486 | No | ||
| 52 | POU4F3 | 2690035 | 10150 | -0.040 | -0.2829 | No | ||
| 53 | MYOZ3 | 1990138 | 10170 | -0.041 | -0.2833 | No | ||
| 54 | SLCO5A1 | 3170040 | 10276 | -0.042 | -0.2883 | No | ||
| 55 | SLC22A8 | 3520204 | 10384 | -0.045 | -0.2933 | No | ||
| 56 | TBX20 | 2230279 4920102 6980020 | 10541 | -0.049 | -0.3010 | No | ||
| 57 | HOXC5 | 1580017 | 10763 | -0.054 | -0.3120 | No | ||
| 58 | TBX2 | 1990563 | 10887 | -0.056 | -0.3177 | No | ||
| 59 | GJA4 | 6380452 | 11066 | -0.060 | -0.3264 | No | ||
| 60 | LECT1 | 2640528 | 11327 | -0.069 | -0.3393 | No | ||
| 61 | UCN | 6620338 | 11656 | -0.079 | -0.3557 | No | ||
| 62 | CNN1 | 510707 | 11727 | -0.082 | -0.3582 | No | ||
| 63 | NFIB | 460450 | 11926 | -0.089 | -0.3674 | No | ||
| 64 | CFL1 | 2340735 | 12117 | -0.097 | -0.3760 | No | ||
| 65 | UBE2E2 | 610053 | 12728 | -0.131 | -0.4068 | No | ||
| 66 | A2BP1 | 2370390 4590593 5550014 | 13078 | -0.157 | -0.4231 | No | ||
| 67 | NR2F1 | 4120528 360402 | 13192 | -0.165 | -0.4265 | No | ||
| 68 | STAT5B | 6200026 | 13637 | -0.210 | -0.4471 | No | ||
| 69 | SGCG | 4920161 | 13713 | -0.217 | -0.4476 | No | ||
| 70 | HSPB7 | 1190441 | 13729 | -0.220 | -0.4448 | No | ||
| 71 | SALL1 | 5420020 7050195 | 13903 | -0.239 | -0.4502 | No | ||
| 72 | IRAK1 | 4120593 | 14010 | -0.252 | -0.4518 | No | ||
| 73 | COL3A1 | 6420273 6620044 | 14059 | -0.260 | -0.4502 | No | ||
| 74 | HMGN2 | 3140091 | 14503 | -0.332 | -0.4687 | No | ||
| 75 | PPAP2B | 3190397 4730280 | 14538 | -0.339 | -0.4650 | No | ||
| 76 | MRGPRF | 4780746 | 14552 | -0.341 | -0.4601 | No | ||
| 77 | HOXA10 | 6110397 7100458 | 14575 | -0.345 | -0.4556 | No | ||
| 78 | KLC2 | 1240048 5340441 | 14724 | -0.375 | -0.4575 | No | ||
| 79 | PPP1R14C | 2570692 5290204 6420184 | 14807 | -0.393 | -0.4555 | No | ||
| 80 | FMNL3 | 3990026 6940577 | 14898 | -0.413 | -0.4536 | No | ||
| 81 | ACTG2 | 4780180 | 15160 | -0.485 | -0.4598 | No | ||
| 82 | POLDIP3 | 670242 1780086 | 15437 | -0.582 | -0.4652 | No | ||
| 83 | GSTP1 | 3170102 3710707 5080368 | 15649 | -0.665 | -0.4657 | No | ||
| 84 | IMPDH2 | 5220138 | 15678 | -0.677 | -0.4562 | No | ||
| 85 | POU3F4 | 870274 | 15748 | -0.707 | -0.4483 | No | ||
| 86 | GRK6 | 1500053 | 16470 | -1.105 | -0.4692 | Yes | ||
| 87 | ARF6 | 3520026 | 16714 | -1.273 | -0.4615 | Yes | ||
| 88 | VASP | 7050500 | 16831 | -1.374 | -0.4453 | Yes | ||
| 89 | FLNA | 5390193 | 17445 | -1.860 | -0.4480 | Yes | ||
| 90 | CNN2 | 2230433 5270446 | 17495 | -1.897 | -0.4196 | Yes | ||
| 91 | SDC1 | 3440471 6350408 | 17881 | -2.353 | -0.4020 | Yes | ||
| 92 | ETHE1 | 3440133 | 17915 | -2.417 | -0.3642 | Yes | ||
| 93 | ETV5 | 110017 | 18034 | -2.603 | -0.3280 | Yes | ||
| 94 | PDLIM7 | 4780136 | 18123 | -2.731 | -0.2882 | Yes | ||
| 95 | MUS81 | 6660184 | 18155 | -2.781 | -0.2444 | Yes | ||
| 96 | SESN3 | 110687 | 18197 | -2.870 | -0.1997 | Yes | ||
| 97 | KLF2 | 6860270 | 18279 | -3.087 | -0.1536 | Yes | ||
| 98 | PNKP | 4610685 5720605 | 18578 | -4.908 | -0.0894 | Yes | ||
| 99 | ALDOA | 6290672 | 18596 | -5.593 | 0.0011 | Yes |

