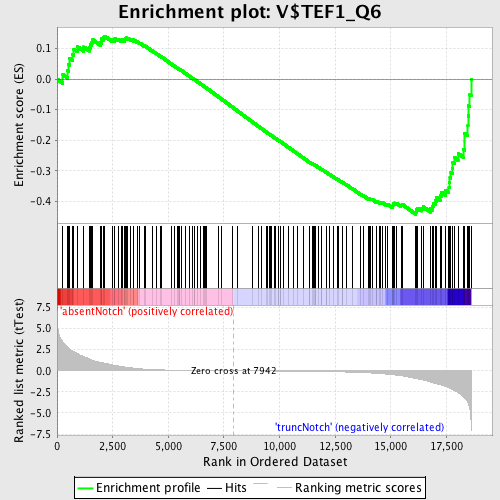
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch_repos |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#absentNotch_versus_truncNotch_repos |
| Upregulated in class | truncNotch |
| GeneSet | V$TEF1_Q6 |
| Enrichment Score (ES) | -0.44304487 |
| Normalized Enrichment Score (NES) | -1.2619562 |
| Nominal p-value | 0.046357617 |
| FDR q-value | 0.8776871 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ITGB1 | 5080156 6270528 | 260 | 3.381 | 0.0151 | No | ||
| 2 | ATP1B1 | 3130594 | 478 | 2.762 | 0.0273 | No | ||
| 3 | PPFIA1 | 430176 3610524 | 511 | 2.694 | 0.0488 | No | ||
| 4 | SLC6A13 | 5270152 | 564 | 2.549 | 0.0680 | No | ||
| 5 | CNN3 | 6110020 | 686 | 2.342 | 0.0817 | No | ||
| 6 | ARID1A | 2630022 1690551 4810110 | 752 | 2.236 | 0.0975 | No | ||
| 7 | VCL | 4120487 | 901 | 2.044 | 0.1071 | No | ||
| 8 | ADIPOR1 | 3140440 | 1202 | 1.664 | 0.1053 | No | ||
| 9 | CDCA3 | 3940397 | 1450 | 1.392 | 0.1039 | No | ||
| 10 | IL18BP | 3450128 | 1499 | 1.342 | 0.1129 | No | ||
| 11 | FXYD6 | 3870164 | 1562 | 1.278 | 0.1206 | No | ||
| 12 | CCND2 | 5340167 | 1608 | 1.231 | 0.1288 | No | ||
| 13 | GATA3 | 6130068 | 1948 | 0.991 | 0.1190 | No | ||
| 14 | MAN1A2 | 5700435 5720114 | 1993 | 0.967 | 0.1250 | No | ||
| 15 | IFRG15 | 2810538 | 2003 | 0.963 | 0.1328 | No | ||
| 16 | LAMC1 | 4730025 | 2095 | 0.908 | 0.1357 | No | ||
| 17 | CDC14A | 4050132 | 2143 | 0.881 | 0.1408 | No | ||
| 18 | ANK3 | 1500161 2900435 4210154 6400100 | 2469 | 0.695 | 0.1292 | No | ||
| 19 | MCAM | 5130270 6620577 | 2597 | 0.635 | 0.1278 | No | ||
| 20 | UBXD3 | 5050193 | 2598 | 0.634 | 0.1333 | No | ||
| 21 | MADD | 6100725 7000088 | 2739 | 0.566 | 0.1306 | No | ||
| 22 | MAST2 | 6200348 | 2905 | 0.496 | 0.1260 | No | ||
| 23 | CHKA | 510324 | 2931 | 0.486 | 0.1288 | No | ||
| 24 | EGR2 | 3800403 | 3028 | 0.452 | 0.1275 | No | ||
| 25 | OVGP1 | 2630746 2940372 | 3037 | 0.448 | 0.1309 | No | ||
| 26 | MTX1 | 5910059 6550181 | 3072 | 0.432 | 0.1328 | No | ||
| 27 | TIMM23 | 60288 7040133 | 3102 | 0.425 | 0.1349 | No | ||
| 28 | SHC1 | 2900731 3170504 6520537 | 3176 | 0.398 | 0.1344 | No | ||
| 29 | ELAVL4 | 50735 3360086 5220167 | 3310 | 0.352 | 0.1303 | No | ||
| 30 | HDGF | 5130114 5420066 6590152 | 3426 | 0.318 | 0.1268 | No | ||
| 31 | PEA15 | 4590685 | 3440 | 0.313 | 0.1288 | No | ||
| 32 | FGFR2 | 4670601 5570402 6940377 | 3600 | 0.265 | 0.1225 | No | ||
| 33 | ABCG4 | 3440458 | 3720 | 0.235 | 0.1181 | No | ||
| 34 | MDK | 3840020 | 3910 | 0.196 | 0.1095 | No | ||
| 35 | OLFML3 | 7100463 | 3959 | 0.186 | 0.1085 | No | ||
| 36 | USP2 | 1190292 1240253 4850035 | 4272 | 0.138 | 0.0928 | No | ||
| 37 | KISS1 | 1580398 | 4457 | 0.117 | 0.0839 | No | ||
| 38 | LAMC2 | 450692 2680041 4200059 4670148 | 4629 | 0.102 | 0.0755 | No | ||
| 39 | WNT3A | 6350348 | 4710 | 0.096 | 0.0720 | No | ||
| 40 | NPPB | 4150722 | 5161 | 0.068 | 0.0482 | No | ||
| 41 | SCNN1A | 4730019 | 5275 | 0.064 | 0.0426 | No | ||
| 42 | NOTCH2 | 2570397 | 5411 | 0.058 | 0.0358 | No | ||
| 43 | EPS8L2 | 2850528 | 5465 | 0.056 | 0.0334 | No | ||
| 44 | KCNIP2 | 60088 1780324 | 5484 | 0.055 | 0.0329 | No | ||
| 45 | LIN28 | 6590672 | 5515 | 0.054 | 0.0318 | No | ||
| 46 | HOOK1 | 840450 940390 2480500 2690402 | 5570 | 0.053 | 0.0293 | No | ||
| 47 | LOXL4 | 6520095 | 5775 | 0.046 | 0.0187 | No | ||
| 48 | LHX4 | 6940551 | 5941 | 0.041 | 0.0101 | No | ||
| 49 | OVOL1 | 1690131 | 6078 | 0.037 | 0.0030 | No | ||
| 50 | POGK | 5290315 | 6154 | 0.035 | -0.0007 | No | ||
| 51 | SDCCAG8 | 1780687 | 6170 | 0.035 | -0.0012 | No | ||
| 52 | DR1 | 5860750 | 6325 | 0.031 | -0.0093 | No | ||
| 53 | PRDM16 | 4120541 | 6425 | 0.029 | -0.0144 | No | ||
| 54 | MPZ | 1990333 | 6582 | 0.025 | -0.0226 | No | ||
| 55 | DNM3 | 730711 | 6633 | 0.024 | -0.0251 | No | ||
| 56 | LAMB3 | 7100301 | 6691 | 0.023 | -0.0280 | No | ||
| 57 | IER5 | 2350575 | 6696 | 0.023 | -0.0280 | No | ||
| 58 | PBX1 | 6660301 | 7248 | 0.012 | -0.0578 | No | ||
| 59 | TLX1 | 2630162 | 7391 | 0.009 | -0.0654 | No | ||
| 60 | KIRREL3 | 2630195 | 7889 | 0.001 | -0.0923 | No | ||
| 61 | RAB18 | 3120575 4590577 | 8119 | -0.003 | -0.1047 | No | ||
| 62 | SYT2 | 60609 | 8773 | -0.014 | -0.1399 | No | ||
| 63 | PTPRJ | 4010707 | 9061 | -0.019 | -0.1553 | No | ||
| 64 | VANGL1 | 380162 | 9190 | -0.021 | -0.1620 | No | ||
| 65 | RNF2 | 2340497 | 9420 | -0.025 | -0.1742 | No | ||
| 66 | MFAP5 | 4920139 | 9448 | -0.025 | -0.1755 | No | ||
| 67 | PRRX1 | 4120193 4480390 | 9563 | -0.028 | -0.1814 | No | ||
| 68 | ATPAF1 | 2470164 | 9572 | -0.028 | -0.1816 | No | ||
| 69 | CGN | 5890139 6370167 6400537 | 9618 | -0.029 | -0.1838 | No | ||
| 70 | SLC1A7 | 1990181 | 9767 | -0.032 | -0.1915 | No | ||
| 71 | POGZ | 870348 7040286 | 9837 | -0.033 | -0.1950 | No | ||
| 72 | TIAL1 | 4150048 6510605 | 9953 | -0.036 | -0.2009 | No | ||
| 73 | BDNF | 2940128 3520368 | 9969 | -0.036 | -0.2014 | No | ||
| 74 | CPT1A | 6350093 | 10027 | -0.038 | -0.2041 | No | ||
| 75 | ANXA8 | 4780022 | 10044 | -0.038 | -0.2047 | No | ||
| 76 | BBOX1 | 2030538 2370452 | 10153 | -0.040 | -0.2102 | No | ||
| 77 | RTN4RL2 | 6450609 | 10404 | -0.045 | -0.2233 | No | ||
| 78 | EIF2C1 | 6900551 | 10611 | -0.050 | -0.2341 | No | ||
| 79 | DAB1 | 2060193 7000605 | 10786 | -0.054 | -0.2430 | No | ||
| 80 | ACTA1 | 840538 | 11083 | -0.061 | -0.2585 | No | ||
| 81 | SYT8 | 4280551 | 11356 | -0.070 | -0.2726 | No | ||
| 82 | THBS3 | 2360465 4050176 | 11362 | -0.070 | -0.2723 | No | ||
| 83 | TNNT2 | 2450364 | 11468 | -0.073 | -0.2774 | No | ||
| 84 | TNRC4 | 4050156 | 11533 | -0.075 | -0.2802 | No | ||
| 85 | ETV3 | 5360487 | 11554 | -0.076 | -0.2806 | No | ||
| 86 | JMJD1C | 940575 2120025 | 11594 | -0.077 | -0.2821 | No | ||
| 87 | FOLR1 | 2470369 | 11630 | -0.078 | -0.2833 | No | ||
| 88 | NES | 1190672 2450167 | 11767 | -0.083 | -0.2899 | No | ||
| 89 | TNNI3K | 5890072 | 11861 | -0.087 | -0.2942 | No | ||
| 90 | ANKRD1 | 4850685 | 11902 | -0.089 | -0.2956 | No | ||
| 91 | CACNA1S | 4730465 | 12107 | -0.097 | -0.3058 | No | ||
| 92 | LMOD1 | 2120035 | 12109 | -0.097 | -0.3050 | No | ||
| 93 | CACNA1E | 840253 | 12261 | -0.104 | -0.3123 | No | ||
| 94 | NNMT | 450471 | 12413 | -0.113 | -0.3195 | No | ||
| 95 | SLC17A6 | 4210576 | 12590 | -0.122 | -0.3280 | No | ||
| 96 | KIF1B | 1240494 2370139 4570270 6510102 | 12650 | -0.126 | -0.3301 | No | ||
| 97 | CYR61 | 1240408 5290026 4120452 6550008 | 12825 | -0.138 | -0.3383 | No | ||
| 98 | JAM3 | 4280193 | 12848 | -0.139 | -0.3383 | No | ||
| 99 | ST5 | 3780204 | 13009 | -0.152 | -0.3457 | No | ||
| 100 | CYP26A1 | 380022 2690600 | 13255 | -0.170 | -0.3575 | No | ||
| 101 | TMPRSS4 | 1050402 1710128 2480341 | 13655 | -0.210 | -0.3773 | No | ||
| 102 | SYTL2 | 580097 5390576 6770603 | 13760 | -0.224 | -0.3809 | No | ||
| 103 | TNKS1BP1 | 7040184 | 13996 | -0.250 | -0.3915 | No | ||
| 104 | ELF3 | 770551 6180019 | 14033 | -0.256 | -0.3912 | No | ||
| 105 | PRKCDBP | 4210341 | 14099 | -0.266 | -0.3925 | No | ||
| 106 | CUL2 | 4200278 | 14153 | -0.274 | -0.3930 | No | ||
| 107 | SYNPO2L | 5390719 | 14360 | -0.308 | -0.4015 | No | ||
| 108 | VAX1 | 6660750 | 14364 | -0.310 | -0.3989 | No | ||
| 109 | HMGN2 | 3140091 | 14503 | -0.332 | -0.4036 | No | ||
| 110 | MRGPRF | 4780746 | 14552 | -0.341 | -0.4032 | No | ||
| 111 | SLC6A9 | 2470520 | 14642 | -0.358 | -0.4049 | No | ||
| 112 | PVRL1 | 2570167 5220519 5890500 3190309 | 14778 | -0.388 | -0.4089 | No | ||
| 113 | EPHA2 | 5890056 | 14854 | -0.403 | -0.4095 | No | ||
| 114 | RAPSN | 610110 2370162 | 15082 | -0.461 | -0.4178 | No | ||
| 115 | CDKN2C | 5050750 5130148 | 15096 | -0.464 | -0.4145 | No | ||
| 116 | SWAP70 | 2810577 2850739 5130594 | 15100 | -0.466 | -0.4106 | No | ||
| 117 | SRRM1 | 6110025 | 15118 | -0.469 | -0.4075 | No | ||
| 118 | MUC1 | 2470273 3140082 6220131 7040066 | 15158 | -0.485 | -0.4054 | No | ||
| 119 | EPC1 | 1570050 5290095 6900193 | 15244 | -0.514 | -0.4056 | No | ||
| 120 | PAQR6 | 580450 2630093 5550341 | 15457 | -0.589 | -0.4120 | No | ||
| 121 | PADI2 | 2940092 6420136 | 15515 | -0.607 | -0.4098 | No | ||
| 122 | PAX2 | 6040270 7000133 | 16130 | -0.916 | -0.4351 | Yes | ||
| 123 | PIAS3 | 50204 240739 4060524 | 16142 | -0.924 | -0.4277 | Yes | ||
| 124 | BRMS1 | 6860280 | 16203 | -0.964 | -0.4227 | Yes | ||
| 125 | EPHB2 | 1090288 1410377 2100541 | 16370 | -1.047 | -0.4226 | Yes | ||
| 126 | CNNM2 | 2650100 | 16454 | -1.096 | -0.4176 | Yes | ||
| 127 | HSPB2 | 1990110 | 16779 | -1.322 | -0.4238 | Yes | ||
| 128 | ADD3 | 4150739 5390600 | 16871 | -1.415 | -0.4165 | Yes | ||
| 129 | PBXIP1 | 70131 | 16904 | -1.442 | -0.4057 | Yes | ||
| 130 | SIPA1 | 5220687 | 17003 | -1.500 | -0.3981 | Yes | ||
| 131 | CRYAB | 4810619 | 17039 | -1.517 | -0.3869 | Yes | ||
| 132 | DPM3 | 50039 | 17215 | -1.643 | -0.3822 | Yes | ||
| 133 | RBM14 | 5340731 5690315 | 17264 | -1.692 | -0.3701 | Yes | ||
| 134 | SH3BGRL3 | 1850750 | 17462 | -1.875 | -0.3646 | Yes | ||
| 135 | EHD1 | 3840731 | 17612 | -2.017 | -0.3552 | Yes | ||
| 136 | AVPI1 | 1850377 | 17633 | -2.039 | -0.3387 | Yes | ||
| 137 | IPO13 | 3870088 | 17657 | -2.058 | -0.3222 | Yes | ||
| 138 | PTPRCAP | 6650075 | 17700 | -2.109 | -0.3062 | Yes | ||
| 139 | DEDD | 6400131 | 17771 | -2.203 | -0.2910 | Yes | ||
| 140 | CHD4 | 5420059 6130338 6380717 | 17789 | -2.234 | -0.2726 | Yes | ||
| 141 | NFKB2 | 2320670 | 17853 | -2.310 | -0.2561 | Yes | ||
| 142 | LMNA | 520471 1500075 3190167 4210020 | 18022 | -2.565 | -0.2430 | Yes | ||
| 143 | RASGRP2 | 2970333 | 18253 | -3.026 | -0.2293 | Yes | ||
| 144 | BCL9L | 6940053 | 18318 | -3.207 | -0.2051 | Yes | ||
| 145 | PTPRF | 1770528 3190044 | 18319 | -3.211 | -0.1773 | Yes | ||
| 146 | USP5 | 4850017 | 18465 | -3.756 | -0.1527 | Yes | ||
| 147 | FRAG1 | 1050504 1170152 2030092 | 18470 | -3.789 | -0.1202 | Yes | ||
| 148 | CD2 | 430672 | 18496 | -3.932 | -0.0876 | Yes | ||
| 149 | POLD4 | 2320082 | 18553 | -4.501 | -0.0517 | Yes | ||
| 150 | MAPKAPK2 | 2850500 | 18609 | -6.376 | 0.0004 | Yes |

