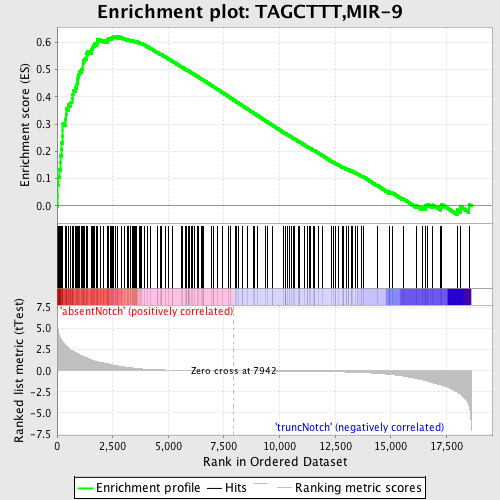
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch_repos |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#absentNotch_versus_truncNotch_repos |
| Upregulated in class | absentNotch |
| GeneSet | TAGCTTT,MIR-9 |
| Enrichment Score (ES) | 0.6232088 |
| Normalized Enrichment Score (NES) | 1.8705649 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0014787808 |
| FWER p-Value | 0.0050 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | GSPT1 | 5420050 | 18 | 5.469 | 0.0376 | Yes | ||
| 2 | RAB14 | 6860139 | 21 | 5.314 | 0.0750 | Yes | ||
| 3 | STK4 | 2640152 | 43 | 4.814 | 0.1078 | Yes | ||
| 4 | BCL11B | 2680673 | 111 | 4.094 | 0.1330 | Yes | ||
| 5 | KPNB1 | 1690138 | 152 | 3.826 | 0.1578 | Yes | ||
| 6 | KRAS | 2060170 | 159 | 3.794 | 0.1843 | Yes | ||
| 7 | MTMR12 | 7000181 | 194 | 3.630 | 0.2080 | Yes | ||
| 8 | PDK1 | 5290427 | 214 | 3.564 | 0.2321 | Yes | ||
| 9 | RAP2C | 1690132 | 251 | 3.418 | 0.2543 | Yes | ||
| 10 | ITGB1 | 5080156 6270528 | 260 | 3.381 | 0.2777 | Yes | ||
| 11 | SENP1 | 6100537 | 261 | 3.381 | 0.3015 | Yes | ||
| 12 | NDEL1 | 2370465 | 380 | 2.993 | 0.3163 | Yes | ||
| 13 | SOCS4 | 1170162 | 399 | 2.942 | 0.3360 | Yes | ||
| 14 | SKP1A | 2450102 | 407 | 2.919 | 0.3562 | Yes | ||
| 15 | DDX3X | 2190020 | 494 | 2.725 | 0.3708 | Yes | ||
| 16 | ETF1 | 6770075 | 620 | 2.447 | 0.3813 | Yes | ||
| 17 | MAP3K7 | 6040068 | 670 | 2.361 | 0.3953 | Yes | ||
| 18 | PSMD7 | 2030619 6220594 | 697 | 2.328 | 0.4103 | Yes | ||
| 19 | EIF4ENIF1 | 2060341 | 719 | 2.292 | 0.4253 | Yes | ||
| 20 | LRP12 | 2370112 | 832 | 2.131 | 0.4343 | Yes | ||
| 21 | YAF2 | 940053 | 890 | 2.061 | 0.4457 | Yes | ||
| 22 | SAR1B | 2510273 | 913 | 2.023 | 0.4588 | Yes | ||
| 23 | SOX4 | 2260091 | 935 | 1.992 | 0.4717 | Yes | ||
| 24 | SFMBT1 | 2360450 2370070 | 976 | 1.918 | 0.4831 | Yes | ||
| 25 | TSGA14 | 6660465 | 1020 | 1.867 | 0.4939 | Yes | ||
| 26 | VAMP4 | 4150102 | 1098 | 1.779 | 0.5023 | Yes | ||
| 27 | APRIN | 2810050 | 1162 | 1.712 | 0.5109 | Yes | ||
| 28 | FBXO33 | 1510504 | 1163 | 1.704 | 0.5229 | Yes | ||
| 29 | FYCO1 | 3990112 | 1173 | 1.694 | 0.5344 | Yes | ||
| 30 | UBE2D3 | 3190452 | 1252 | 1.609 | 0.5415 | Yes | ||
| 31 | MIB1 | 3800537 | 1327 | 1.521 | 0.5482 | Yes | ||
| 32 | CUGBP2 | 6180121 6840139 | 1328 | 1.521 | 0.5590 | Yes | ||
| 33 | PHF2 | 4060300 | 1383 | 1.466 | 0.5664 | Yes | ||
| 34 | ARNTL | 3170463 | 1538 | 1.301 | 0.5672 | Yes | ||
| 35 | YWHAZ | 1230717 | 1558 | 1.283 | 0.5752 | Yes | ||
| 36 | CCND2 | 5340167 | 1608 | 1.231 | 0.5813 | Yes | ||
| 37 | NFKBIZ | 2970450 | 1615 | 1.228 | 0.5896 | Yes | ||
| 38 | TNFRSF19L | 1170168 1190358 5670270 | 1677 | 1.165 | 0.5945 | Yes | ||
| 39 | PPP3R1 | 1190041 1570487 | 1772 | 1.092 | 0.5971 | Yes | ||
| 40 | MECP2 | 1940450 | 1803 | 1.075 | 0.6031 | Yes | ||
| 41 | ELOVL6 | 5340746 | 1825 | 1.064 | 0.6094 | Yes | ||
| 42 | LRP4 | 1740253 | 1945 | 0.993 | 0.6100 | Yes | ||
| 43 | KCTD9 | 780577 | 2105 | 0.903 | 0.6077 | Yes | ||
| 44 | KCNN2 | 990025 3520524 | 2259 | 0.811 | 0.6051 | Yes | ||
| 45 | NR3C1 | 630563 | 2279 | 0.804 | 0.6098 | Yes | ||
| 46 | HIPK1 | 110193 | 2313 | 0.786 | 0.6135 | Yes | ||
| 47 | TIMP3 | 1450504 1980270 | 2396 | 0.745 | 0.6143 | Yes | ||
| 48 | PCAF | 2230161 2570369 6550451 | 2450 | 0.708 | 0.6165 | Yes | ||
| 49 | ZHX1 | 2100110 2120286 | 2503 | 0.674 | 0.6184 | Yes | ||
| 50 | TRIM33 | 580619 2230280 3990433 6200747 | 2532 | 0.659 | 0.6215 | Yes | ||
| 51 | PHF16 | 50373 7200161 | 2612 | 0.627 | 0.6217 | Yes | ||
| 52 | STC1 | 360161 | 2728 | 0.571 | 0.6195 | Yes | ||
| 53 | SORCS1 | 60411 5890373 | 2734 | 0.570 | 0.6232 | Yes | ||
| 54 | GSPT2 | 6590674 | 2909 | 0.495 | 0.6173 | No | ||
| 55 | STRN3 | 1450093 6200685 | 3021 | 0.454 | 0.6144 | No | ||
| 56 | ANKRD10 | 1240025 | 3175 | 0.399 | 0.6090 | No | ||
| 57 | CYGB | 870347 | 3204 | 0.389 | 0.6102 | No | ||
| 58 | E2F3 | 50162 460180 | 3320 | 0.350 | 0.6064 | No | ||
| 59 | KRIT1 | 2640500 4230435 4730184 | 3403 | 0.328 | 0.6043 | No | ||
| 60 | DNAJC6 | 4590438 5900070 | 3415 | 0.323 | 0.6060 | No | ||
| 61 | NLK | 2030010 2450041 | 3499 | 0.295 | 0.6036 | No | ||
| 62 | ZFP36L1 | 2510138 4120048 | 3513 | 0.292 | 0.6049 | No | ||
| 63 | CASK | 2340215 3290576 6770215 | 3581 | 0.271 | 0.6032 | No | ||
| 64 | RBBP5 | 2760735 6760164 | 3688 | 0.242 | 0.5992 | No | ||
| 65 | SLITRK2 | 2640020 | 3769 | 0.222 | 0.5964 | No | ||
| 66 | PPP1R8 | 6290292 | 3785 | 0.219 | 0.5971 | No | ||
| 67 | ERBB2IP | 580253 1090672 | 3909 | 0.196 | 0.5918 | No | ||
| 68 | SP5 | 3840309 | 4080 | 0.164 | 0.5838 | No | ||
| 69 | ATP11A | 1240717 5570603 | 4182 | 0.150 | 0.5794 | No | ||
| 70 | NR4A3 | 2900021 5860095 5910039 | 4516 | 0.112 | 0.5621 | No | ||
| 71 | E2F7 | 4200056 | 4524 | 0.111 | 0.5625 | No | ||
| 72 | AKT3 | 1580270 3290278 | 4634 | 0.102 | 0.5573 | No | ||
| 73 | PLCG1 | 6020369 | 4678 | 0.098 | 0.5557 | No | ||
| 74 | PSCD3 | 730301 | 4685 | 0.098 | 0.5560 | No | ||
| 75 | HMGB2 | 2640603 | 4856 | 0.085 | 0.5474 | No | ||
| 76 | SOX21 | 1050110 4670041 6660338 | 5023 | 0.075 | 0.5389 | No | ||
| 77 | QKI | 5220093 6130044 | 5165 | 0.068 | 0.5318 | No | ||
| 78 | SMARCA1 | 4230315 | 5177 | 0.067 | 0.5316 | No | ||
| 79 | TLL2 | 6220040 | 5593 | 0.052 | 0.5095 | No | ||
| 80 | RARG | 6760136 | 5614 | 0.051 | 0.5088 | No | ||
| 81 | OSBPL8 | 50603 | 5750 | 0.047 | 0.5018 | No | ||
| 82 | GOLGA4 | 460300 | 5816 | 0.045 | 0.4986 | No | ||
| 83 | MEF2C | 670025 780338 | 5821 | 0.044 | 0.4987 | No | ||
| 84 | DCX | 130075 5360025 7050673 | 5883 | 0.043 | 0.4957 | No | ||
| 85 | HNRPH2 | 2630040 5720671 | 5895 | 0.042 | 0.4954 | No | ||
| 86 | PRPF4B | 940181 2570300 | 5905 | 0.042 | 0.4952 | No | ||
| 87 | TMEM47 | 1660086 | 5939 | 0.041 | 0.4937 | No | ||
| 88 | ARHGAP5 | 2510619 3360035 | 6037 | 0.038 | 0.4887 | No | ||
| 89 | LRRC4 | 3850041 | 6083 | 0.037 | 0.4865 | No | ||
| 90 | EDG1 | 4200619 | 6152 | 0.035 | 0.4831 | No | ||
| 91 | DR1 | 5860750 | 6325 | 0.031 | 0.4740 | No | ||
| 92 | SET | 6650286 | 6363 | 0.030 | 0.4722 | No | ||
| 93 | PRKCE | 5700053 | 6487 | 0.027 | 0.4657 | No | ||
| 94 | MITF | 380056 | 6523 | 0.026 | 0.4640 | No | ||
| 95 | PCDH18 | 430010 | 6562 | 0.026 | 0.4621 | No | ||
| 96 | SLITRK6 | 7320739 | 6601 | 0.025 | 0.4602 | No | ||
| 97 | SLC16A1 | 50433 | 6944 | 0.018 | 0.4418 | No | ||
| 98 | TEAD1 | 2470551 | 7024 | 0.016 | 0.4377 | No | ||
| 99 | HS6ST2 | 520133 5550603 | 7046 | 0.016 | 0.4366 | No | ||
| 100 | TCEA1 | 5340070 | 7201 | 0.013 | 0.4284 | No | ||
| 101 | CFL2 | 380239 6200368 | 7413 | 0.009 | 0.4170 | No | ||
| 102 | CRK | 1230162 4780128 | 7694 | 0.004 | 0.4018 | No | ||
| 103 | PCTK2 | 5700673 | 7791 | 0.002 | 0.3966 | No | ||
| 104 | NR2F2 | 3170609 3310577 | 8001 | -0.001 | 0.3853 | No | ||
| 105 | ZIC2 | 1410408 | 8056 | -0.002 | 0.3824 | No | ||
| 106 | FBXO22 | 4760010 | 8134 | -0.003 | 0.3783 | No | ||
| 107 | ZFPM2 | 460072 | 8346 | -0.007 | 0.3669 | No | ||
| 108 | MMP16 | 2680139 | 8350 | -0.007 | 0.3667 | No | ||
| 109 | CRISPLD1 | 5910524 | 8568 | -0.010 | 0.3551 | No | ||
| 110 | PARD6B | 4120600 | 8848 | -0.015 | 0.3400 | No | ||
| 111 | GABRB3 | 4150164 | 8891 | -0.016 | 0.3379 | No | ||
| 112 | THRB | 1780673 | 8997 | -0.017 | 0.3323 | No | ||
| 113 | SLITRK3 | 5910041 | 9363 | -0.024 | 0.3127 | No | ||
| 114 | DVL2 | 6110162 | 9445 | -0.025 | 0.3085 | No | ||
| 115 | WAC | 5340390 5910164 | 9660 | -0.030 | 0.2971 | No | ||
| 116 | INSM1 | 3450671 | 9698 | -0.030 | 0.2953 | No | ||
| 117 | SOX3 | 4570537 | 10159 | -0.040 | 0.2706 | No | ||
| 118 | LRRTM1 | 1690324 | 10255 | -0.042 | 0.2658 | No | ||
| 119 | KCNJ3 | 50184 | 10283 | -0.043 | 0.2646 | No | ||
| 120 | PI15 | 3840133 | 10285 | -0.043 | 0.2648 | No | ||
| 121 | FRAS1 | 5080170 5670347 | 10371 | -0.045 | 0.2606 | No | ||
| 122 | GABRA1 | 130372 | 10443 | -0.046 | 0.2570 | No | ||
| 123 | AFF3 | 3870102 | 10524 | -0.048 | 0.2530 | No | ||
| 124 | NEGR1 | 1940731 5130170 | 10626 | -0.050 | 0.2479 | No | ||
| 125 | CIT | 2370601 | 10658 | -0.051 | 0.2466 | No | ||
| 126 | CNTN4 | 1780300 5570577 6370019 | 10830 | -0.055 | 0.2377 | No | ||
| 127 | DPYSL3 | 1580594 | 10877 | -0.056 | 0.2356 | No | ||
| 128 | AKAP2 | 5550347 | 10895 | -0.056 | 0.2351 | No | ||
| 129 | MPP5 | 2100148 5220632 | 11117 | -0.062 | 0.2235 | No | ||
| 130 | HTR2C | 380497 | 11242 | -0.065 | 0.2173 | No | ||
| 131 | CPEB3 | 3940164 | 11250 | -0.066 | 0.2174 | No | ||
| 132 | POU4F1 | 2260315 | 11335 | -0.069 | 0.2133 | No | ||
| 133 | GPM6A | 1660044 2750152 | 11392 | -0.071 | 0.2108 | No | ||
| 134 | SLC9A9 | 1940309 | 11538 | -0.075 | 0.2034 | No | ||
| 135 | ACTL6A | 5220543 6290541 2450093 4590332 5270722 | 11564 | -0.076 | 0.2026 | No | ||
| 136 | CALD1 | 1770129 1940397 | 11583 | -0.077 | 0.2022 | No | ||
| 137 | THBD | 1500092 | 11761 | -0.083 | 0.1932 | No | ||
| 138 | NAP1L5 | 360576 | 11946 | -0.090 | 0.1838 | No | ||
| 139 | ATXN1 | 5550156 | 12348 | -0.109 | 0.1628 | No | ||
| 140 | HOXB5 | 2900538 | 12437 | -0.114 | 0.1589 | No | ||
| 141 | ZFHX4 | 6110167 | 12510 | -0.118 | 0.1558 | No | ||
| 142 | NDST1 | 1500427 5340121 | 12525 | -0.119 | 0.1559 | No | ||
| 143 | SLC25A37 | 6520341 460471 | 12634 | -0.125 | 0.1509 | No | ||
| 144 | CYR61 | 1240408 5290026 4120452 6550008 | 12825 | -0.138 | 0.1416 | No | ||
| 145 | SOX2 | 630487 2230491 | 12863 | -0.141 | 0.1406 | No | ||
| 146 | TPD52 | 670463 4610541 | 12873 | -0.142 | 0.1411 | No | ||
| 147 | HOXB13 | 4730152 | 12987 | -0.150 | 0.1360 | No | ||
| 148 | STX16 | 70315 | 13017 | -0.152 | 0.1355 | No | ||
| 149 | ENAH | 1690292 5700300 | 13104 | -0.158 | 0.1320 | No | ||
| 150 | ELAVL2 | 360181 | 13114 | -0.159 | 0.1326 | No | ||
| 151 | SORCS3 | 5700309 | 13221 | -0.167 | 0.1280 | No | ||
| 152 | EPHA7 | 1190288 4010278 | 13222 | -0.167 | 0.1292 | No | ||
| 153 | POU3F2 | 6620484 | 13280 | -0.172 | 0.1273 | No | ||
| 154 | HIP2 | 2810095 3990369 4120301 | 13428 | -0.184 | 0.1207 | No | ||
| 155 | CREB5 | 2320368 | 13496 | -0.191 | 0.1184 | No | ||
| 156 | GPHN | 3290195 3840121 | 13698 | -0.216 | 0.1090 | No | ||
| 157 | BACH2 | 6760131 | 13765 | -0.225 | 0.1070 | No | ||
| 158 | PHF6 | 3120632 3990041 | 14389 | -0.313 | 0.0754 | No | ||
| 159 | CPEB2 | 4760338 | 14942 | -0.423 | 0.0485 | No | ||
| 160 | MAGI1 | 6510500 1990239 4670471 | 14947 | -0.425 | 0.0513 | No | ||
| 161 | STAT1 | 6510204 6590553 | 15078 | -0.460 | 0.0475 | No | ||
| 162 | CAMK2G | 630097 | 15552 | -0.623 | 0.0262 | No | ||
| 163 | PIGF | 5690050 | 16157 | -0.937 | 0.0001 | No | ||
| 164 | TARDBP | 870390 2350093 | 16405 | -1.070 | -0.0058 | No | ||
| 165 | HNRPA2B1 | 1240619 6200161 6200494 6980408 | 16546 | -1.160 | -0.0052 | No | ||
| 166 | SERF2 | 5720010 | 16563 | -1.169 | 0.0022 | No | ||
| 167 | SFRS2 | 50707 380593 | 16661 | -1.237 | 0.0057 | No | ||
| 168 | ZMYND11 | 630181 2570019 4850138 5360040 | 16891 | -1.433 | 0.0034 | No | ||
| 169 | NCKIPSD | 2940156 | 17237 | -1.668 | -0.0036 | No | ||
| 170 | TPM1 | 130673 | 17289 | -1.716 | 0.0058 | No | ||
| 171 | HRB | 1660397 6110398 | 17978 | -2.507 | -0.0138 | No | ||
| 172 | PITPNM2 | 1580044 | 18144 | -2.766 | -0.0033 | No | ||
| 173 | DTX3 | 1690458 | 18513 | -4.087 | 0.0056 | No |

