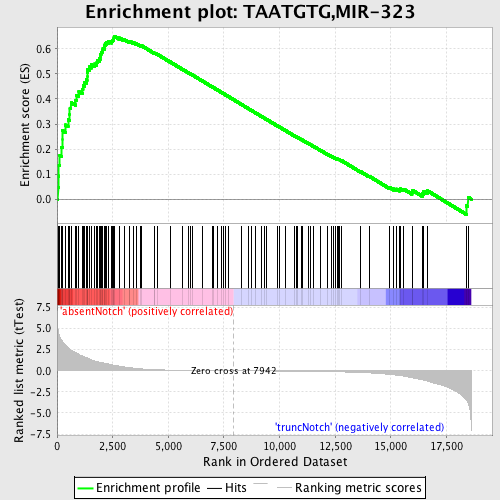
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch_repos |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#absentNotch_versus_truncNotch_repos |
| Upregulated in class | absentNotch |
| GeneSet | TAATGTG,MIR-323 |
| Enrichment Score (ES) | 0.6507365 |
| Normalized Enrichment Score (NES) | 1.8590193 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0013402553 |
| FWER p-Value | 0.0060 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SFRS1 | 2360440 | 39 | 4.846 | 0.0466 | Yes | ||
| 2 | SATB1 | 5670154 | 55 | 4.668 | 0.0927 | Yes | ||
| 3 | TMPO | 4050494 | 74 | 4.448 | 0.1364 | Yes | ||
| 4 | MAPRE1 | 3290037 | 122 | 4.031 | 0.1743 | Yes | ||
| 5 | RANBP9 | 4670685 | 187 | 3.680 | 0.2078 | Yes | ||
| 6 | ARFIP1 | 3130601 | 250 | 3.423 | 0.2389 | Yes | ||
| 7 | RAB1A | 2370671 | 252 | 3.405 | 0.2730 | Yes | ||
| 8 | NDEL1 | 2370465 | 380 | 2.993 | 0.2962 | Yes | ||
| 9 | DDX3X | 2190020 | 494 | 2.725 | 0.3175 | Yes | ||
| 10 | DHX36 | 2470465 | 572 | 2.528 | 0.3388 | Yes | ||
| 11 | PITPNA | 2760332 | 578 | 2.518 | 0.3638 | Yes | ||
| 12 | UBL3 | 6450458 | 624 | 2.441 | 0.3859 | Yes | ||
| 13 | KPNA1 | 5270324 | 839 | 2.121 | 0.3956 | Yes | ||
| 14 | SMARCAD1 | 5390619 | 874 | 2.084 | 0.4147 | Yes | ||
| 15 | CEPT1 | 1940047 | 979 | 1.916 | 0.4283 | Yes | ||
| 16 | UTX | 2900017 6770452 | 1130 | 1.741 | 0.4377 | Yes | ||
| 17 | FEM1B | 1230110 | 1164 | 1.704 | 0.4531 | Yes | ||
| 18 | CUL1 | 1990632 | 1241 | 1.620 | 0.4652 | Yes | ||
| 19 | CSNK1G3 | 110450 | 1322 | 1.527 | 0.4762 | Yes | ||
| 20 | NFIX | 2450152 | 1346 | 1.505 | 0.4901 | Yes | ||
| 21 | HS2ST1 | 3170088 | 1350 | 1.497 | 0.5050 | Yes | ||
| 22 | ZDHHC21 | 2370338 | 1374 | 1.475 | 0.5186 | Yes | ||
| 23 | SUMO1 | 1990364 2760338 6380195 | 1458 | 1.382 | 0.5280 | Yes | ||
| 24 | PJA2 | 430537 | 1544 | 1.296 | 0.5364 | Yes | ||
| 25 | GJA1 | 5220731 | 1700 | 1.150 | 0.5396 | Yes | ||
| 26 | MRC1 | 730097 | 1789 | 1.085 | 0.5457 | Yes | ||
| 27 | REV3L | 1090717 | 1823 | 1.065 | 0.5546 | Yes | ||
| 28 | SLMAP | 3120093 4760128 5390528 | 1897 | 1.014 | 0.5609 | Yes | ||
| 29 | TNRC6A | 1240537 6130037 6200010 | 1934 | 0.998 | 0.5690 | Yes | ||
| 30 | WBP2 | 3170128 | 1953 | 0.988 | 0.5779 | Yes | ||
| 31 | ENTPD5 | 4280070 | 1976 | 0.978 | 0.5866 | Yes | ||
| 32 | ULK2 | 50041 5050176 | 2024 | 0.953 | 0.5936 | Yes | ||
| 33 | MAP4K4 | 2360059 | 2060 | 0.933 | 0.6011 | Yes | ||
| 34 | CHCHD4 | 2680075 | 2121 | 0.892 | 0.6068 | Yes | ||
| 35 | RPS6KA3 | 1980707 | 2130 | 0.889 | 0.6153 | Yes | ||
| 36 | CLASP1 | 6860279 | 2182 | 0.859 | 0.6212 | Yes | ||
| 37 | KPNA3 | 7040088 | 2227 | 0.834 | 0.6272 | Yes | ||
| 38 | HIPK1 | 110193 | 2313 | 0.786 | 0.6305 | Yes | ||
| 39 | PDCD10 | 4760500 | 2451 | 0.708 | 0.6302 | Yes | ||
| 40 | ZHX1 | 2100110 2120286 | 2503 | 0.674 | 0.6342 | Yes | ||
| 41 | TRIM33 | 580619 2230280 3990433 6200747 | 2532 | 0.659 | 0.6393 | Yes | ||
| 42 | ASF1A | 7050020 | 2556 | 0.651 | 0.6446 | Yes | ||
| 43 | RAB10 | 2360008 5890020 | 2564 | 0.648 | 0.6507 | Yes | ||
| 44 | NFAT5 | 2510411 5890195 6550152 | 2801 | 0.540 | 0.6434 | No | ||
| 45 | ATP11B | 4230100 | 3022 | 0.454 | 0.6361 | No | ||
| 46 | DCUN1D1 | 2360332 | 3254 | 0.375 | 0.6274 | No | ||
| 47 | TOP2B | 6980309 | 3262 | 0.372 | 0.6307 | No | ||
| 48 | CREB1 | 1500717 2230358 3610600 6550601 | 3414 | 0.323 | 0.6258 | No | ||
| 49 | HOOK3 | 1190242 2470059 | 3554 | 0.279 | 0.6211 | No | ||
| 50 | SLITRK2 | 2640020 | 3769 | 0.222 | 0.6118 | No | ||
| 51 | ANKRD27 | 2190465 4070300 6040026 | 3814 | 0.214 | 0.6115 | No | ||
| 52 | PCDH9 | 2100136 | 4371 | 0.126 | 0.5828 | No | ||
| 53 | PPARGC1A | 4670040 | 4383 | 0.125 | 0.5834 | No | ||
| 54 | NR4A3 | 2900021 5860095 5910039 | 4516 | 0.112 | 0.5774 | No | ||
| 55 | TBPL1 | 3610332 | 5075 | 0.073 | 0.5480 | No | ||
| 56 | RSBN1 | 7000487 | 5617 | 0.051 | 0.5193 | No | ||
| 57 | POU4F2 | 2120195 2570022 | 5886 | 0.042 | 0.5052 | No | ||
| 58 | FGD4 | 520168 870411 2640253 6550338 6650364 | 6015 | 0.039 | 0.4987 | No | ||
| 59 | LRRC4 | 3850041 | 6083 | 0.037 | 0.4955 | No | ||
| 60 | WTAP | 4010176 | 6552 | 0.026 | 0.4704 | No | ||
| 61 | HOXA1 | 1190524 5420142 | 6970 | 0.017 | 0.4481 | No | ||
| 62 | TEAD1 | 2470551 | 7024 | 0.016 | 0.4454 | No | ||
| 63 | AUH | 5570152 | 7200 | 0.013 | 0.4361 | No | ||
| 64 | TGFA | 1570332 | 7388 | 0.009 | 0.4260 | No | ||
| 65 | QSCN6L1 | 870040 | 7485 | 0.008 | 0.4209 | No | ||
| 66 | EBF3 | 4200129 | 7590 | 0.006 | 0.4154 | No | ||
| 67 | CRK | 1230162 4780128 | 7694 | 0.004 | 0.4099 | No | ||
| 68 | BET1 | 4010725 | 8281 | -0.006 | 0.3782 | No | ||
| 69 | RCN1 | 2480041 | 8611 | -0.011 | 0.3606 | No | ||
| 70 | MCFD2 | 520576 5690017 | 8750 | -0.013 | 0.3533 | No | ||
| 71 | NRK | 4590609 | 8894 | -0.016 | 0.3457 | No | ||
| 72 | MYLIP | 50717 4050672 | 9195 | -0.021 | 0.3297 | No | ||
| 73 | CLCN3 | 510195 780176 1660093 3830372 | 9333 | -0.023 | 0.3225 | No | ||
| 74 | STX12 | 610451 | 9431 | -0.025 | 0.3175 | No | ||
| 75 | LPP | 1660541 | 9890 | -0.034 | 0.2931 | No | ||
| 76 | DPF2 | 6980373 | 9998 | -0.037 | 0.2877 | No | ||
| 77 | KCNJ3 | 50184 | 10283 | -0.043 | 0.2728 | No | ||
| 78 | SEMA6D | 4050324 5860138 6350307 | 10682 | -0.052 | 0.2518 | No | ||
| 79 | FBN2 | 50435 | 10774 | -0.054 | 0.2475 | No | ||
| 80 | EIF3S1 | 6130368 6770044 | 10825 | -0.055 | 0.2453 | No | ||
| 81 | PAK7 | 3830164 | 10980 | -0.058 | 0.2376 | No | ||
| 82 | SEPT3 | 3830022 5290079 | 11012 | -0.059 | 0.2365 | No | ||
| 83 | ZIC1 | 670113 | 11283 | -0.067 | 0.2226 | No | ||
| 84 | GPM6A | 1660044 2750152 | 11392 | -0.071 | 0.2174 | No | ||
| 85 | STK35 | 2360121 | 11527 | -0.075 | 0.2110 | No | ||
| 86 | TMOD1 | 3850100 | 11860 | -0.087 | 0.1939 | No | ||
| 87 | RAB11FIP2 | 4570040 | 12137 | -0.098 | 0.1800 | No | ||
| 88 | SLC4A4 | 1780075 1850040 4810068 | 12334 | -0.109 | 0.1705 | No | ||
| 89 | HOXB5 | 2900538 | 12437 | -0.114 | 0.1661 | No | ||
| 90 | PKP4 | 3710400 5890097 | 12495 | -0.117 | 0.1642 | No | ||
| 91 | POU2F3 | 2030601 | 12584 | -0.122 | 0.1607 | No | ||
| 92 | CXCL12 | 580546 4150750 4570068 | 12660 | -0.127 | 0.1579 | No | ||
| 93 | USP9X | 130435 2370735 3120338 | 12681 | -0.128 | 0.1581 | No | ||
| 94 | OSBP | 5390471 | 12793 | -0.136 | 0.1535 | No | ||
| 95 | STAT5B | 6200026 | 13637 | -0.210 | 0.1100 | No | ||
| 96 | UHRF2 | 60050 840280 1340746 5080091 | 14021 | -0.254 | 0.0919 | No | ||
| 97 | CCND1 | 460524 770309 3120576 6980398 | 14927 | -0.419 | 0.0472 | No | ||
| 98 | PPP1CB | 3130411 3780270 4150180 | 15130 | -0.473 | 0.0410 | No | ||
| 99 | CDKN1B | 3800025 6450044 | 15233 | -0.512 | 0.0407 | No | ||
| 100 | NFKB1 | 5420358 | 15387 | -0.564 | 0.0381 | No | ||
| 101 | EGR3 | 6940128 | 15440 | -0.582 | 0.0411 | No | ||
| 102 | ZNF318 | 5890091 6130021 | 15585 | -0.636 | 0.0397 | No | ||
| 103 | TMEM16D | 2650324 | 15952 | -0.819 | 0.0282 | No | ||
| 104 | TOP1 | 770471 4060632 6650324 | 15975 | -0.831 | 0.0353 | No | ||
| 105 | FBN1 | 3170181 | 16435 | -1.085 | 0.0214 | No | ||
| 106 | MAP3K3 | 610685 | 16480 | -1.118 | 0.0303 | No | ||
| 107 | ACAT2 | 2680400 3610440 | 16636 | -1.222 | 0.0342 | No | ||
| 108 | PTK2B | 4730411 | 18400 | -3.448 | -0.0264 | No | ||
| 109 | SPSB4 | 4070673 | 18468 | -3.787 | 0.0080 | No |

