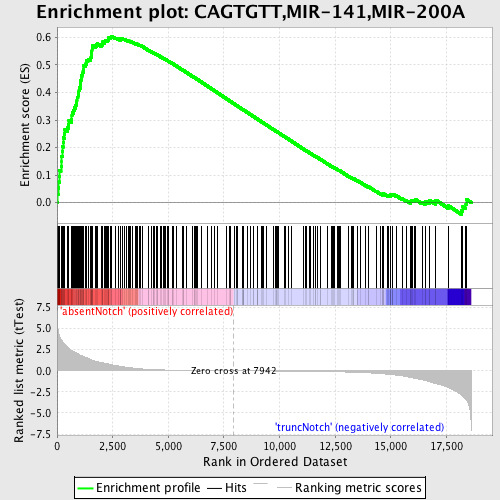
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch_repos |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#absentNotch_versus_truncNotch_repos |
| Upregulated in class | absentNotch |
| GeneSet | CAGTGTT,MIR-141,MIR-200A |
| Enrichment Score (ES) | 0.6045511 |
| Normalized Enrichment Score (NES) | 1.8724961 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0017745368 |
| FWER p-Value | 0.0050 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PRKACB | 4210170 | 9 | 6.062 | 0.0313 | Yes | ||
| 2 | WSB1 | 870563 | 47 | 4.783 | 0.0543 | Yes | ||
| 3 | ANP32E | 6510706 | 81 | 4.345 | 0.0752 | Yes | ||
| 4 | SUPT6H | 3170082 | 96 | 4.172 | 0.0963 | Yes | ||
| 5 | PPP2CA | 3990113 | 115 | 4.067 | 0.1167 | Yes | ||
| 6 | MAP2K4 | 5130133 | 175 | 3.735 | 0.1330 | Yes | ||
| 7 | PITPNB | 5130039 | 203 | 3.593 | 0.1504 | Yes | ||
| 8 | TRAM1 | 2340441 6620592 | 207 | 3.588 | 0.1690 | Yes | ||
| 9 | SFPQ | 4760110 | 235 | 3.477 | 0.1857 | Yes | ||
| 10 | RAP2C | 1690132 | 251 | 3.418 | 0.2028 | Yes | ||
| 11 | CD47 | 1850603 | 270 | 3.343 | 0.2193 | Yes | ||
| 12 | RNF44 | 5910692 | 290 | 3.259 | 0.2354 | Yes | ||
| 13 | RABGAP1 | 5050397 | 326 | 3.128 | 0.2498 | Yes | ||
| 14 | OSBPL11 | 1770044 | 348 | 3.068 | 0.2648 | Yes | ||
| 15 | ATP1B1 | 3130594 | 478 | 2.762 | 0.2722 | Yes | ||
| 16 | MTPN | 3940167 | 515 | 2.686 | 0.2843 | Yes | ||
| 17 | CCNE2 | 3120537 | 529 | 2.639 | 0.2974 | Yes | ||
| 18 | CNP | 2260451 | 648 | 2.398 | 0.3036 | Yes | ||
| 19 | ATP6V1A | 6590242 | 666 | 2.364 | 0.3150 | Yes | ||
| 20 | H2AFZ | 1470168 | 673 | 2.360 | 0.3271 | Yes | ||
| 21 | BRD3 | 6130672 | 742 | 2.246 | 0.3351 | Yes | ||
| 22 | YTHDF2 | 4060332 4610279 | 772 | 2.212 | 0.3451 | Yes | ||
| 23 | PPP1R15B | 2640280 5860082 | 817 | 2.153 | 0.3540 | Yes | ||
| 24 | SLC20A1 | 6420014 | 855 | 2.104 | 0.3630 | Yes | ||
| 25 | YAF2 | 940053 | 890 | 2.061 | 0.3720 | Yes | ||
| 26 | CDV3 | 1940577 | 894 | 2.057 | 0.3826 | Yes | ||
| 27 | ATP2A2 | 1090075 3990279 | 939 | 1.987 | 0.3906 | Yes | ||
| 28 | RBMS1 | 6400014 | 954 | 1.950 | 0.4001 | Yes | ||
| 29 | GRB2 | 6650398 | 982 | 1.909 | 0.4086 | Yes | ||
| 30 | GPC2 | 130647 | 1003 | 1.884 | 0.4174 | Yes | ||
| 31 | CUL3 | 1850520 | 1037 | 1.848 | 0.4252 | Yes | ||
| 32 | RUNX1 | 3840711 | 1043 | 1.837 | 0.4346 | Yes | ||
| 33 | FOXP1 | 6510433 | 1060 | 1.822 | 0.4433 | Yes | ||
| 34 | CBL | 6380068 | 1079 | 1.799 | 0.4517 | Yes | ||
| 35 | SLC23A2 | 6660722 | 1083 | 1.795 | 0.4609 | Yes | ||
| 36 | TULP4 | 2320364 | 1120 | 1.755 | 0.4682 | Yes | ||
| 37 | APRIN | 2810050 | 1162 | 1.712 | 0.4749 | Yes | ||
| 38 | SIRT1 | 1190731 | 1190 | 1.676 | 0.4822 | Yes | ||
| 39 | CDC25A | 3800184 | 1196 | 1.670 | 0.4907 | Yes | ||
| 40 | RTF1 | 2100735 | 1197 | 1.669 | 0.4994 | Yes | ||
| 41 | TCF12 | 3610324 7000156 | 1276 | 1.578 | 0.5034 | Yes | ||
| 42 | CSNK1G3 | 110450 | 1322 | 1.527 | 0.5090 | Yes | ||
| 43 | MIB1 | 3800537 | 1327 | 1.521 | 0.5167 | Yes | ||
| 44 | SIM2 | 2100044 | 1389 | 1.457 | 0.5211 | Yes | ||
| 45 | KIF3A | 5050348 6650079 | 1505 | 1.335 | 0.5218 | Yes | ||
| 46 | DNAJC13 | 4730091 | 1520 | 1.317 | 0.5279 | Yes | ||
| 47 | ARNTL | 3170463 | 1538 | 1.301 | 0.5338 | Yes | ||
| 48 | OGT | 2360131 4610333 | 1542 | 1.298 | 0.5404 | Yes | ||
| 49 | HOXB4 | 540131 | 1545 | 1.296 | 0.5471 | Yes | ||
| 50 | TFRC | 4050551 | 1567 | 1.269 | 0.5526 | Yes | ||
| 51 | ATP6V1B2 | 4060528 | 1569 | 1.268 | 0.5592 | Yes | ||
| 52 | MBNL1 | 2640762 7100048 | 1576 | 1.263 | 0.5655 | Yes | ||
| 53 | YWHAG | 3780341 | 1591 | 1.251 | 0.5713 | Yes | ||
| 54 | OLIG3 | 1050603 | 1719 | 1.140 | 0.5704 | Yes | ||
| 55 | AK3L1 | 7100601 | 1779 | 1.087 | 0.5728 | Yes | ||
| 56 | MECP2 | 1940450 | 1803 | 1.075 | 0.5772 | Yes | ||
| 57 | PDCD4 | 3360403 5690110 | 1981 | 0.974 | 0.5727 | Yes | ||
| 58 | ACACA | 2490612 2680369 | 2022 | 0.954 | 0.5755 | Yes | ||
| 59 | ANKFY1 | 360309 5390021 | 2037 | 0.947 | 0.5797 | Yes | ||
| 60 | FUS | 4760619 | 2058 | 0.935 | 0.5835 | Yes | ||
| 61 | CDC14A | 4050132 | 2143 | 0.881 | 0.5836 | Yes | ||
| 62 | BHLHB2 | 7040603 | 2152 | 0.878 | 0.5877 | Yes | ||
| 63 | KPNA3 | 7040088 | 2227 | 0.834 | 0.5881 | Yes | ||
| 64 | PLAGL2 | 1770148 | 2270 | 0.808 | 0.5900 | Yes | ||
| 65 | SLC39A9 | 3940348 | 2295 | 0.796 | 0.5929 | Yes | ||
| 66 | HIPK1 | 110193 | 2313 | 0.786 | 0.5961 | Yes | ||
| 67 | CALU | 2680403 3520056 4610348 5720176 6130121 | 2324 | 0.779 | 0.5996 | Yes | ||
| 68 | ARHGEF18 | 2120279 | 2389 | 0.749 | 0.6001 | Yes | ||
| 69 | MYH10 | 2640673 | 2426 | 0.718 | 0.6019 | Yes | ||
| 70 | PTGES3 | 2190440 2680524 | 2446 | 0.709 | 0.6046 | Yes | ||
| 71 | TCERG1 | 2350551 4540397 5220746 | 2638 | 0.615 | 0.5974 | No | ||
| 72 | FKBP5 | 2190048 | 2750 | 0.562 | 0.5943 | No | ||
| 73 | THRAP2 | 4920600 | 2843 | 0.524 | 0.5920 | No | ||
| 74 | TNS3 | 6380242 | 2852 | 0.520 | 0.5943 | No | ||
| 75 | YTHDF3 | 4540632 4670717 7040609 | 2866 | 0.515 | 0.5963 | No | ||
| 76 | FBXL2 | 6450722 | 2937 | 0.484 | 0.5950 | No | ||
| 77 | TAF12 | 3710746 4920300 6180603 | 3010 | 0.460 | 0.5935 | No | ||
| 78 | CHD9 | 430037 2570129 5420398 | 3117 | 0.419 | 0.5900 | No | ||
| 79 | MARCH7 | 3800020 6620168 | 3224 | 0.383 | 0.5862 | No | ||
| 80 | CNR1 | 6550494 6760497 | 3248 | 0.377 | 0.5869 | No | ||
| 81 | E2F3 | 50162 460180 | 3320 | 0.350 | 0.5849 | No | ||
| 82 | BICD2 | 1090411 | 3393 | 0.330 | 0.5827 | No | ||
| 83 | SOX11 | 610279 | 3534 | 0.284 | 0.5766 | No | ||
| 84 | WDR44 | 130195 1050609 1570685 | 3565 | 0.276 | 0.5764 | No | ||
| 85 | FGFR2 | 4670601 5570402 6940377 | 3600 | 0.265 | 0.5759 | No | ||
| 86 | RPH3AL | 70110 | 3714 | 0.237 | 0.5710 | No | ||
| 87 | GATA6 | 1770010 | 3747 | 0.228 | 0.5705 | No | ||
| 88 | SLC1A1 | 520372 6420059 | 3839 | 0.210 | 0.5666 | No | ||
| 89 | SPAG9 | 580497 2510180 4210685 | 4123 | 0.157 | 0.5521 | No | ||
| 90 | BAP1 | 3830131 | 4125 | 0.157 | 0.5529 | No | ||
| 91 | SNAP91 | 3170056 5690520 | 4235 | 0.142 | 0.5477 | No | ||
| 92 | PTP4A1 | 130692 | 4343 | 0.130 | 0.5425 | No | ||
| 93 | APBB2 | 5130403 | 4352 | 0.129 | 0.5428 | No | ||
| 94 | PCDH9 | 2100136 | 4371 | 0.126 | 0.5425 | No | ||
| 95 | CSNK1A1 | 2340427 | 4478 | 0.115 | 0.5373 | No | ||
| 96 | DUSP3 | 1240129 6840215 | 4501 | 0.113 | 0.5367 | No | ||
| 97 | STRN | 5570706 7050333 | 4513 | 0.112 | 0.5367 | No | ||
| 98 | STXBP1 | 360072 6220717 | 4668 | 0.099 | 0.5288 | No | ||
| 99 | DRD2 | 5890369 | 4713 | 0.096 | 0.5269 | No | ||
| 100 | GRIN3A | 2370592 | 4803 | 0.089 | 0.5226 | No | ||
| 101 | PBEF1 | 1090278 | 4823 | 0.088 | 0.5220 | No | ||
| 102 | UBE3A | 1240152 2690438 5860609 | 4870 | 0.084 | 0.5199 | No | ||
| 103 | WDR26 | 1090138 | 4945 | 0.080 | 0.5163 | No | ||
| 104 | JUN | 840170 | 4993 | 0.077 | 0.5142 | No | ||
| 105 | KCNJ2 | 630019 | 5013 | 0.076 | 0.5135 | No | ||
| 106 | QKI | 5220093 6130044 | 5165 | 0.068 | 0.5057 | No | ||
| 107 | MYRIP | 1580471 | 5206 | 0.066 | 0.5038 | No | ||
| 108 | RAD23B | 2190671 | 5234 | 0.065 | 0.5027 | No | ||
| 109 | ATXN7 | 1450435 | 5347 | 0.061 | 0.4969 | No | ||
| 110 | ESRRG | 4010181 | 5648 | 0.050 | 0.4809 | No | ||
| 111 | RBM7 | 5890348 | 5692 | 0.049 | 0.4788 | No | ||
| 112 | SEPT7 | 2760685 | 5696 | 0.049 | 0.4789 | No | ||
| 113 | CSNK2A1 | 1580577 | 5807 | 0.045 | 0.4732 | No | ||
| 114 | OLFM1 | 3360161 5570066 | 6075 | 0.038 | 0.4588 | No | ||
| 115 | NR2C2 | 6100097 | 6162 | 0.035 | 0.4544 | No | ||
| 116 | TGFB2 | 4920292 | 6193 | 0.034 | 0.4529 | No | ||
| 117 | DMWD | 770019 1690047 | 6240 | 0.033 | 0.4506 | No | ||
| 118 | ARPC5 | 1740411 3450300 | 6256 | 0.033 | 0.4499 | No | ||
| 119 | DR1 | 5860750 | 6325 | 0.031 | 0.4464 | No | ||
| 120 | SLC16A7 | 2320672 | 6469 | 0.028 | 0.4388 | No | ||
| 121 | HNRPF | 670138 | 6739 | 0.022 | 0.4242 | No | ||
| 122 | CLASP2 | 2510139 | 6949 | 0.018 | 0.4130 | No | ||
| 123 | SNF1LK | 6110403 | 7064 | 0.016 | 0.4069 | No | ||
| 124 | CALCR | 1690494 | 7203 | 0.013 | 0.3994 | No | ||
| 125 | EXOC5 | 7000711 | 7608 | 0.006 | 0.3775 | No | ||
| 126 | SRC | 580132 | 7737 | 0.003 | 0.3705 | No | ||
| 127 | NRXN1 | 460408 5340270 6370066 | 7772 | 0.003 | 0.3687 | No | ||
| 128 | PCTK2 | 5700673 | 7791 | 0.002 | 0.3677 | No | ||
| 129 | LENG8 | 3130731 | 7980 | -0.001 | 0.3575 | No | ||
| 130 | ZFR | 5910647 | 8057 | -0.002 | 0.3534 | No | ||
| 131 | TTR | 4150072 | 8085 | -0.002 | 0.3519 | No | ||
| 132 | SP4 | 3850176 | 8094 | -0.002 | 0.3515 | No | ||
| 133 | MMP16 | 2680139 | 8350 | -0.007 | 0.3377 | No | ||
| 134 | B3GNT1 | 2350446 | 8352 | -0.007 | 0.3377 | No | ||
| 135 | DDX5 | 940021 2630338 3440148 | 8359 | -0.007 | 0.3374 | No | ||
| 136 | HOXA11 | 6980133 | 8363 | -0.007 | 0.3373 | No | ||
| 137 | SPG3A | 2760091 3120170 | 8389 | -0.008 | 0.3359 | No | ||
| 138 | PCDH8 | 2360047 4200452 | 8560 | -0.010 | 0.3268 | No | ||
| 139 | GPM6B | 6220458 | 8670 | -0.012 | 0.3209 | No | ||
| 140 | TRHDE | 1690068 | 8814 | -0.015 | 0.3132 | No | ||
| 141 | LYPLA1 | 4480373 5570551 | 8817 | -0.015 | 0.3132 | No | ||
| 142 | CHES1 | 4570121 5700162 | 8993 | -0.017 | 0.3037 | No | ||
| 143 | THRB | 1780673 | 8997 | -0.017 | 0.3037 | No | ||
| 144 | RARB | 430139 1410138 | 9020 | -0.018 | 0.3026 | No | ||
| 145 | MTSS1 | 780435 2370114 | 9197 | -0.021 | 0.2931 | No | ||
| 146 | ITSN1 | 2640577 | 9240 | -0.022 | 0.2909 | No | ||
| 147 | CSF3 | 2230193 6660707 | 9275 | -0.022 | 0.2892 | No | ||
| 148 | PAFAH1B2 | 5360546 6860672 | 9424 | -0.025 | 0.2813 | No | ||
| 149 | HLF | 2370113 | 9743 | -0.031 | 0.2642 | No | ||
| 150 | FOXA1 | 3710609 | 9825 | -0.033 | 0.2599 | No | ||
| 151 | WDFY3 | 3610041 4560333 4570273 | 9869 | -0.034 | 0.2578 | No | ||
| 152 | NFASC | 1940619 2470156 4590168 6840524 | 9879 | -0.034 | 0.2575 | No | ||
| 153 | LPP | 1660541 | 9890 | -0.034 | 0.2571 | No | ||
| 154 | TIAL1 | 4150048 6510605 | 9953 | -0.036 | 0.2539 | No | ||
| 155 | ACVR2A | 6110647 | 10239 | -0.042 | 0.2386 | No | ||
| 156 | KHDRBS2 | 3520717 | 10252 | -0.042 | 0.2382 | No | ||
| 157 | DIXDC1 | 6980435 | 10381 | -0.045 | 0.2315 | No | ||
| 158 | HOXC13 | 4070722 | 10554 | -0.049 | 0.2224 | No | ||
| 159 | TMEFF1 | 2680100 | 11095 | -0.061 | 0.1933 | No | ||
| 160 | PITX2 | 870537 2690139 | 11144 | -0.063 | 0.1911 | No | ||
| 161 | STAT4 | 1660373 3130019 | 11229 | -0.065 | 0.1868 | No | ||
| 162 | POU4F1 | 2260315 | 11335 | -0.069 | 0.1815 | No | ||
| 163 | PHYHIPL | 2360706 3840692 | 11397 | -0.071 | 0.1785 | No | ||
| 164 | STK35 | 2360121 | 11527 | -0.075 | 0.1719 | No | ||
| 165 | NRP2 | 4070400 5860041 6650446 | 11605 | -0.077 | 0.1681 | No | ||
| 166 | PHLPP | 3440563 | 11629 | -0.078 | 0.1673 | No | ||
| 167 | BRUNOL6 | 6840373 | 11701 | -0.081 | 0.1639 | No | ||
| 168 | KLHL18 | 4560075 | 11833 | -0.086 | 0.1572 | No | ||
| 169 | MYT1L | 5670369 6660403 | 11841 | -0.086 | 0.1572 | No | ||
| 170 | LHX6 | 2360731 3360397 | 12143 | -0.099 | 0.1414 | No | ||
| 171 | ATXN1 | 5550156 | 12348 | -0.109 | 0.1309 | No | ||
| 172 | FRMD4A | 2190176 | 12384 | -0.111 | 0.1296 | No | ||
| 173 | HOXB5 | 2900538 | 12437 | -0.114 | 0.1273 | No | ||
| 174 | CPD | 5360403 5860685 | 12478 | -0.116 | 0.1258 | No | ||
| 175 | SLC17A6 | 4210576 | 12590 | -0.122 | 0.1204 | No | ||
| 176 | CXCL12 | 580546 4150750 4570068 | 12660 | -0.127 | 0.1173 | No | ||
| 177 | USP9X | 130435 2370735 3120338 | 12681 | -0.128 | 0.1169 | No | ||
| 178 | RAB38 | 2970735 | 12726 | -0.131 | 0.1152 | No | ||
| 179 | ELAVL2 | 360181 | 13114 | -0.159 | 0.0950 | No | ||
| 180 | EPHA7 | 1190288 4010278 | 13222 | -0.167 | 0.0900 | No | ||
| 181 | ELF2 | 1690014 2480132 3060180 3190504 | 13296 | -0.173 | 0.0869 | No | ||
| 182 | CYP26B1 | 2630142 | 13319 | -0.175 | 0.0867 | No | ||
| 183 | MDGA1 | 1170139 | 13340 | -0.177 | 0.0865 | No | ||
| 184 | SOX5 | 2370576 2900167 3190128 5050528 | 13506 | -0.192 | 0.0785 | No | ||
| 185 | PLAG1 | 1450142 3870139 6520039 | 13519 | -0.194 | 0.0789 | No | ||
| 186 | STAT5B | 6200026 | 13637 | -0.210 | 0.0736 | No | ||
| 187 | PTPRG | 2650524 1340022 | 13882 | -0.237 | 0.0616 | No | ||
| 188 | TTYH2 | 1450397 3870100 6550347 | 13978 | -0.249 | 0.0577 | No | ||
| 189 | TM9SF4 | 450538 1050093 4210112 | 14000 | -0.251 | 0.0579 | No | ||
| 190 | ZCCHC3 | 580438 1990035 4780278 | 14348 | -0.306 | 0.0407 | No | ||
| 191 | KIF1C | 2480484 | 14556 | -0.341 | 0.0312 | No | ||
| 192 | SYT4 | 5080193 | 14627 | -0.355 | 0.0292 | No | ||
| 193 | SLC6A9 | 2470520 | 14642 | -0.358 | 0.0304 | No | ||
| 194 | SEMA6A | 2690138 3060021 | 14658 | -0.359 | 0.0314 | No | ||
| 195 | PAPPA | 4230463 | 14662 | -0.361 | 0.0331 | No | ||
| 196 | EPHA2 | 5890056 | 14854 | -0.403 | 0.0249 | No | ||
| 197 | HIC2 | 4010433 | 14915 | -0.418 | 0.0238 | No | ||
| 198 | SIPA1L2 | 4850731 | 14971 | -0.431 | 0.0231 | No | ||
| 199 | PEX5 | 1240746 1570092 2510487 | 14989 | -0.435 | 0.0244 | No | ||
| 200 | SPRYD3 | 50403 | 14990 | -0.435 | 0.0267 | No | ||
| 201 | MOBKL2B | 940128 | 14999 | -0.436 | 0.0285 | No | ||
| 202 | DOLPP1 | 4780619 6400053 | 15063 | -0.457 | 0.0275 | No | ||
| 203 | TSC1 | 1850672 | 15080 | -0.461 | 0.0291 | No | ||
| 204 | LRRC8A | 2510193 | 15091 | -0.463 | 0.0309 | No | ||
| 205 | DLC1 | 1090632 6450594 | 15250 | -0.516 | 0.0250 | No | ||
| 206 | CDC42BPB | 6350037 | 15510 | -0.605 | 0.0141 | No | ||
| 207 | HMG20A | 4590113 | 15692 | -0.680 | 0.0079 | No | ||
| 208 | PPP2R2A | 2900014 5700575 | 15890 | -0.786 | 0.0013 | No | ||
| 209 | PPT2 | 2340059 3450148 | 15897 | -0.789 | 0.0051 | No | ||
| 210 | MDM1 | 510528 1780280 2680152 2690181 3060025 3120487 4210601 4540037 4610142 5290110 6400433 6480743 | 15940 | -0.810 | 0.0070 | No | ||
| 211 | TOP1 | 770471 4060632 6650324 | 15975 | -0.831 | 0.0095 | No | ||
| 212 | AMPD2 | 630093 1170070 | 16082 | -0.886 | 0.0084 | No | ||
| 213 | DEK | 1050451 4560037 | 16111 | -0.906 | 0.0116 | No | ||
| 214 | HRBL | 1850102 3830253 4780176 | 16401 | -1.067 | 0.0015 | No | ||
| 215 | ATF5 | 5670685 | 16541 | -1.156 | -0.0000 | No | ||
| 216 | SERF2 | 5720010 | 16563 | -1.169 | 0.0050 | No | ||
| 217 | SLC25A3 | 670112 | 16732 | -1.291 | 0.0026 | No | ||
| 218 | SON | 3120148 5860239 7040403 | 16751 | -1.303 | 0.0084 | No | ||
| 219 | EPN1 | 4060725 | 17017 | -1.507 | 0.0019 | No | ||
| 220 | ST3GAL3 | 2810601 | 17022 | -1.509 | 0.0096 | No | ||
| 221 | BACE1 | 2650438 | 17577 | -1.977 | -0.0102 | No | ||
| 222 | RTN4RL1 | 2630368 | 18181 | -2.848 | -0.0280 | No | ||
| 223 | PSEN1 | 130403 2030647 6100603 | 18233 | -2.978 | -0.0152 | No | ||
| 224 | ACOT7 | 730735 1400176 7050053 | 18375 | -3.382 | -0.0052 | No | ||
| 225 | STAT5A | 2680458 | 18406 | -3.478 | 0.0114 | No |

