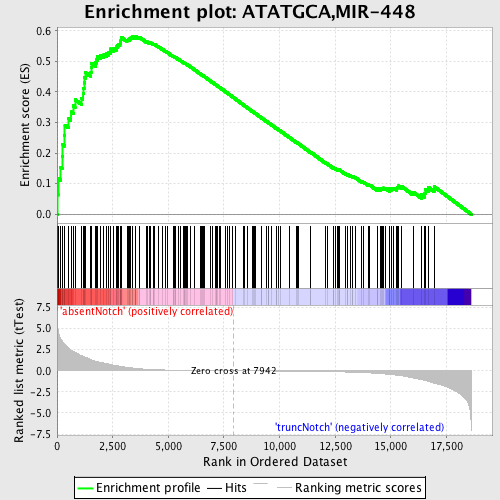
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch_repos |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#absentNotch_versus_truncNotch_repos |
| Upregulated in class | absentNotch |
| GeneSet | ATATGCA,MIR-448 |
| Enrichment Score (ES) | 0.5824525 |
| Normalized Enrichment Score (NES) | 1.732569 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.006328996 |
| FWER p-Value | 0.057 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | APPBP2 | 5130215 | 13 | 5.776 | 0.0646 | Yes | ||
| 2 | STK4 | 2640152 | 43 | 4.814 | 0.1174 | Yes | ||
| 3 | KRAS | 2060170 | 159 | 3.794 | 0.1540 | Yes | ||
| 4 | TOPORS | 4060592 | 227 | 3.500 | 0.1899 | Yes | ||
| 5 | RAP2C | 1690132 | 251 | 3.418 | 0.2273 | Yes | ||
| 6 | KLF13 | 4670563 | 331 | 3.109 | 0.2581 | Yes | ||
| 7 | PELI1 | 3870215 6900040 | 353 | 3.055 | 0.2915 | Yes | ||
| 8 | BRD1 | 1660278 | 522 | 2.668 | 0.3126 | Yes | ||
| 9 | UBL3 | 6450458 | 624 | 2.441 | 0.3347 | Yes | ||
| 10 | DNAJB11 | 4150168 | 731 | 2.270 | 0.3546 | Yes | ||
| 11 | SCN5A | 4070017 | 809 | 2.160 | 0.3748 | Yes | ||
| 12 | AMMECR1 | 3800435 | 1092 | 1.786 | 0.3797 | Yes | ||
| 13 | FBXO33 | 1510504 | 1163 | 1.704 | 0.3952 | Yes | ||
| 14 | SIRT1 | 1190731 | 1190 | 1.676 | 0.4127 | Yes | ||
| 15 | SLC7A11 | 2850138 | 1215 | 1.655 | 0.4301 | Yes | ||
| 16 | CENTG2 | 4210053 | 1236 | 1.626 | 0.4474 | Yes | ||
| 17 | FBXO28 | 1340358 4730100 | 1278 | 1.576 | 0.4630 | Yes | ||
| 18 | PPM2C | 3360209 | 1516 | 1.321 | 0.4651 | Yes | ||
| 19 | RNF19 | 6400315 | 1529 | 1.309 | 0.4792 | Yes | ||
| 20 | ARNTL | 3170463 | 1538 | 1.301 | 0.4935 | Yes | ||
| 21 | LRRIQ2 | 3990064 6370673 6380372 | 1732 | 1.130 | 0.4958 | Yes | ||
| 22 | MAP2K6 | 1230056 2940204 | 1761 | 1.102 | 0.5067 | Yes | ||
| 23 | TPP2 | 3850093 | 1819 | 1.066 | 0.5157 | Yes | ||
| 24 | PCDHGC5 | 2190056 3450504 | 1938 | 0.995 | 0.5205 | Yes | ||
| 25 | KCTD9 | 780577 | 2105 | 0.903 | 0.5218 | Yes | ||
| 26 | KPNA3 | 7040088 | 2227 | 0.834 | 0.5246 | Yes | ||
| 27 | WDR22 | 6200739 | 2306 | 0.793 | 0.5294 | Yes | ||
| 28 | RNGTT | 780152 780746 1090471 | 2390 | 0.749 | 0.5333 | Yes | ||
| 29 | MYH3 | 110400 | 2412 | 0.732 | 0.5405 | Yes | ||
| 30 | DYNLT3 | 5570398 | 2543 | 0.656 | 0.5408 | Yes | ||
| 31 | ATXN3 | 50156 2900095 | 2654 | 0.611 | 0.5418 | Yes | ||
| 32 | MBNL2 | 3450707 | 2679 | 0.596 | 0.5472 | Yes | ||
| 33 | PRKAR2B | 3130593 5220577 | 2715 | 0.577 | 0.5518 | Yes | ||
| 34 | DPP4 | 5720369 | 2752 | 0.561 | 0.5562 | Yes | ||
| 35 | DLGAP4 | 4570672 5690161 | 2827 | 0.529 | 0.5582 | Yes | ||
| 36 | THRAP2 | 4920600 | 2843 | 0.524 | 0.5633 | Yes | ||
| 37 | YTHDF3 | 4540632 4670717 7040609 | 2866 | 0.515 | 0.5679 | Yes | ||
| 38 | KLHL5 | 3060484 6400292 | 2871 | 0.511 | 0.5735 | Yes | ||
| 39 | NTF3 | 5890131 | 2897 | 0.500 | 0.5778 | Yes | ||
| 40 | FUSIP1 | 520082 5390114 | 3143 | 0.410 | 0.5691 | Yes | ||
| 41 | KCTD15 | 2810735 | 3191 | 0.395 | 0.5711 | Yes | ||
| 42 | LYST | 3870039 7000717 | 3256 | 0.374 | 0.5718 | Yes | ||
| 43 | NRIP1 | 3190039 | 3269 | 0.369 | 0.5753 | Yes | ||
| 44 | E2F3 | 50162 460180 | 3320 | 0.350 | 0.5766 | Yes | ||
| 45 | SMURF1 | 520609 580647 | 3380 | 0.334 | 0.5772 | Yes | ||
| 46 | BICD2 | 1090411 | 3393 | 0.330 | 0.5802 | Yes | ||
| 47 | PCDHGB1 | 3060504 1340338 | 3523 | 0.289 | 0.5765 | Yes | ||
| 48 | NCOA2 | 940347 2850725 | 3532 | 0.285 | 0.5793 | Yes | ||
| 49 | SOX11 | 610279 | 3534 | 0.284 | 0.5825 | Yes | ||
| 50 | RANBP2 | 4280338 | 3686 | 0.242 | 0.5770 | No | ||
| 51 | PCDHGB2 | 5130408 | 3721 | 0.234 | 0.5778 | No | ||
| 52 | DTNA | 1340600 1780731 2340278 2850132 | 4019 | 0.174 | 0.5637 | No | ||
| 53 | PRKAB2 | 6980300 | 4059 | 0.167 | 0.5635 | No | ||
| 54 | KLF5 | 3840348 | 4156 | 0.153 | 0.5600 | No | ||
| 55 | ATP11A | 1240717 5570603 | 4182 | 0.150 | 0.5603 | No | ||
| 56 | PCDHGB6 | 2570433 | 4199 | 0.148 | 0.5611 | No | ||
| 57 | DACT1 | 1500050 | 4335 | 0.131 | 0.5553 | No | ||
| 58 | APBB2 | 5130403 | 4352 | 0.129 | 0.5559 | No | ||
| 59 | PCDH9 | 2100136 | 4371 | 0.126 | 0.5563 | No | ||
| 60 | DBC1 | 4610156 | 4573 | 0.107 | 0.5467 | No | ||
| 61 | DAPK1 | 5570010 | 4744 | 0.093 | 0.5385 | No | ||
| 62 | ZCCHC2 | 460592 | 4866 | 0.085 | 0.5329 | No | ||
| 63 | AZIN1 | 3450070 | 4942 | 0.081 | 0.5297 | No | ||
| 64 | SP3 | 3840338 | 5216 | 0.066 | 0.5157 | No | ||
| 65 | GNAI1 | 4560390 | 5227 | 0.065 | 0.5159 | No | ||
| 66 | RORB | 6400035 | 5290 | 0.063 | 0.5133 | No | ||
| 67 | PCDHGA10 | 2450138 | 5301 | 0.063 | 0.5134 | No | ||
| 68 | RAB2B | 6200091 | 5306 | 0.062 | 0.5139 | No | ||
| 69 | RAB21 | 5570070 | 5475 | 0.056 | 0.5054 | No | ||
| 70 | PCDHGA8 | 2120593 | 5536 | 0.054 | 0.5028 | No | ||
| 71 | KCNB1 | 3610035 | 5676 | 0.049 | 0.4958 | No | ||
| 72 | AFF2 | 1410487 | 5711 | 0.048 | 0.4945 | No | ||
| 73 | IGF1R | 3360494 | 5719 | 0.048 | 0.4947 | No | ||
| 74 | PCDHGA1 | 5050372 | 5758 | 0.046 | 0.4931 | No | ||
| 75 | MEF2C | 670025 780338 | 5821 | 0.044 | 0.4903 | No | ||
| 76 | TMSB4X | 6620114 | 5876 | 0.043 | 0.4878 | No | ||
| 77 | PCDHGA4 | 6100035 | 5978 | 0.040 | 0.4828 | No | ||
| 78 | BNC2 | 4810603 | 6167 | 0.035 | 0.4730 | No | ||
| 79 | ADD1 | 1500176 3060139 3390092 | 6456 | 0.028 | 0.4578 | No | ||
| 80 | ETV1 | 70735 2940603 5080463 | 6483 | 0.027 | 0.4567 | No | ||
| 81 | WTAP | 4010176 | 6552 | 0.026 | 0.4533 | No | ||
| 82 | KCNMB2 | 3780128 4760136 | 6594 | 0.025 | 0.4513 | No | ||
| 83 | DNM3 | 730711 | 6633 | 0.024 | 0.4495 | No | ||
| 84 | PCDHGB5 | 6350577 | 6903 | 0.018 | 0.4352 | No | ||
| 85 | NLGN1 | 5670278 | 6987 | 0.017 | 0.4309 | No | ||
| 86 | SYN2 | 3990593 | 7098 | 0.015 | 0.4251 | No | ||
| 87 | PFN2 | 730040 5700026 | 7174 | 0.014 | 0.4212 | No | ||
| 88 | BRP44L | 1340605 | 7185 | 0.013 | 0.4208 | No | ||
| 89 | BTG3 | 7050079 | 7193 | 0.013 | 0.4205 | No | ||
| 90 | PCDHGA9 | 460292 | 7293 | 0.011 | 0.4153 | No | ||
| 91 | ADAM10 | 3780156 | 7346 | 0.010 | 0.4126 | No | ||
| 92 | CRY1 | 6770039 | 7568 | 0.006 | 0.4007 | No | ||
| 93 | SERTAD2 | 2810673 6840450 | 7658 | 0.005 | 0.3959 | No | ||
| 94 | TCEAL1 | 870577 | 7744 | 0.003 | 0.3914 | No | ||
| 95 | PCDHGA5 | 2260750 | 7881 | 0.001 | 0.3840 | No | ||
| 96 | CASP4 | 5860047 6650093 | 7997 | -0.001 | 0.3778 | No | ||
| 97 | HOXA11 | 6980133 | 8363 | -0.007 | 0.3581 | No | ||
| 98 | CLCN5 | 4200390 | 8433 | -0.008 | 0.3544 | No | ||
| 99 | PCDH8 | 2360047 4200452 | 8560 | -0.010 | 0.3477 | No | ||
| 100 | GLULD1 | 5690592 | 8800 | -0.014 | 0.3349 | No | ||
| 101 | FBXW11 | 6450632 | 8838 | -0.015 | 0.3331 | No | ||
| 102 | GABRB3 | 4150164 | 8891 | -0.016 | 0.3305 | No | ||
| 103 | SH3GL2 | 1990176 3830494 | 8909 | -0.016 | 0.3297 | No | ||
| 104 | PCDHGB4 | 3830292 | 9166 | -0.021 | 0.3161 | No | ||
| 105 | PURA | 3870156 | 9192 | -0.021 | 0.3150 | No | ||
| 106 | WRNIP1 | 2320541 5890725 | 9428 | -0.025 | 0.3025 | No | ||
| 107 | COLEC12 | 2690717 4590086 | 9503 | -0.027 | 0.2988 | No | ||
| 108 | ANK2 | 6510546 | 9507 | -0.027 | 0.2989 | No | ||
| 109 | SS18 | 1940440 2900072 6900129 | 9629 | -0.029 | 0.2927 | No | ||
| 110 | WDFY3 | 3610041 4560333 4570273 | 9869 | -0.034 | 0.2802 | No | ||
| 111 | PCDHGA2 | 1780541 | 9878 | -0.034 | 0.2801 | No | ||
| 112 | PCDHGA6 | 3390242 | 9966 | -0.036 | 0.2758 | No | ||
| 113 | FMR1 | 5050075 | 10022 | -0.037 | 0.2732 | No | ||
| 114 | PCDHGA3 | 1570112 | 10437 | -0.046 | 0.2513 | No | ||
| 115 | PLEKHA6 | 6040041 7040270 | 10760 | -0.054 | 0.2345 | No | ||
| 116 | NAV1 | 4210519 6620129 | 10787 | -0.054 | 0.2337 | No | ||
| 117 | CNTN4 | 1780300 5570577 6370019 | 10830 | -0.055 | 0.2320 | No | ||
| 118 | DOC2A | 6020121 | 11376 | -0.070 | 0.2033 | No | ||
| 119 | GPM6A | 1660044 2750152 | 11392 | -0.071 | 0.2033 | No | ||
| 120 | GRIK2 | 4610164 | 12047 | -0.094 | 0.1689 | No | ||
| 121 | RAB11FIP2 | 4570040 | 12137 | -0.098 | 0.1652 | No | ||
| 122 | EPHA4 | 460750 | 12418 | -0.113 | 0.1513 | No | ||
| 123 | GDF6 | 1340048 | 12435 | -0.114 | 0.1518 | No | ||
| 124 | ZFHX4 | 6110167 | 12510 | -0.118 | 0.1491 | No | ||
| 125 | PCDHGB7 | 460309 | 12614 | -0.124 | 0.1449 | No | ||
| 126 | CXCL12 | 580546 4150750 4570068 | 12660 | -0.127 | 0.1439 | No | ||
| 127 | AHR | 4560671 | 12673 | -0.128 | 0.1447 | No | ||
| 128 | TBX3 | 2570672 | 12682 | -0.128 | 0.1457 | No | ||
| 129 | CACNA1C | 1660128 3780300 3870092 6290403 6900301 | 12940 | -0.146 | 0.1334 | No | ||
| 130 | CLK1 | 1780551 2100102 2340347 2480347 | 13056 | -0.155 | 0.1289 | No | ||
| 131 | VLDLR | 870722 3060047 5340452 6550131 | 13073 | -0.156 | 0.1298 | No | ||
| 132 | ELL2 | 6420091 | 13189 | -0.165 | 0.1255 | No | ||
| 133 | ARMC8 | 520408 940132 4060368 4210102 | 13291 | -0.173 | 0.1220 | No | ||
| 134 | ELF2 | 1690014 2480132 3060180 3190504 | 13296 | -0.173 | 0.1237 | No | ||
| 135 | PCDHGC4 | 2900152 | 13391 | -0.180 | 0.1206 | No | ||
| 136 | GPHN | 3290195 3840121 | 13698 | -0.216 | 0.1065 | No | ||
| 137 | BACH2 | 6760131 | 13765 | -0.225 | 0.1055 | No | ||
| 138 | SF3B1 | 7000632 | 13982 | -0.249 | 0.0966 | No | ||
| 139 | PCDHGA7 | 2350025 | 14055 | -0.259 | 0.0956 | No | ||
| 140 | PTBP1 | 4070736 | 14401 | -0.315 | 0.0805 | No | ||
| 141 | HECTD2 | 2680014 3060689 | 14414 | -0.318 | 0.0834 | No | ||
| 142 | SOCS5 | 3830398 7100093 | 14544 | -0.339 | 0.0803 | No | ||
| 143 | UNC5A | 2120528 3060324 | 14561 | -0.342 | 0.0833 | No | ||
| 144 | BCL11A | 6860369 | 14617 | -0.352 | 0.0843 | No | ||
| 145 | PAPPA | 4230463 | 14662 | -0.361 | 0.0859 | No | ||
| 146 | ING3 | 110438 2060390 | 14752 | -0.380 | 0.0854 | No | ||
| 147 | CPEB2 | 4760338 | 14942 | -0.423 | 0.0800 | No | ||
| 148 | AZI2 | 1400300 2680463 | 14951 | -0.426 | 0.0843 | No | ||
| 149 | OTX2 | 1190471 | 15050 | -0.453 | 0.0842 | No | ||
| 150 | BCL2 | 730132 1570736 2470138 3800044 4810037 5690068 5860504 6650164 | 15135 | -0.474 | 0.0850 | No | ||
| 151 | KCNH7 | 7510068 | 15267 | -0.522 | 0.0838 | No | ||
| 152 | DOCK9 | 5080037 | 15282 | -0.527 | 0.0890 | No | ||
| 153 | SH3KBP1 | 670102 2850458 7000309 | 15327 | -0.542 | 0.0927 | No | ||
| 154 | CAP1 | 2650278 | 15489 | -0.600 | 0.0908 | No | ||
| 155 | GLCE | 4850040 | 16000 | -0.845 | 0.0727 | No | ||
| 156 | EPHB2 | 1090288 1410377 2100541 | 16370 | -1.047 | 0.0645 | No | ||
| 157 | FBXL17 | 770170 | 16527 | -1.148 | 0.0690 | No | ||
| 158 | HNRPA2B1 | 1240619 6200161 6200494 6980408 | 16546 | -1.160 | 0.0812 | No | ||
| 159 | XYLT2 | 6380039 | 16700 | -1.267 | 0.0872 | No | ||
| 160 | ARID2 | 110647 5390435 | 16945 | -1.467 | 0.0905 | No |