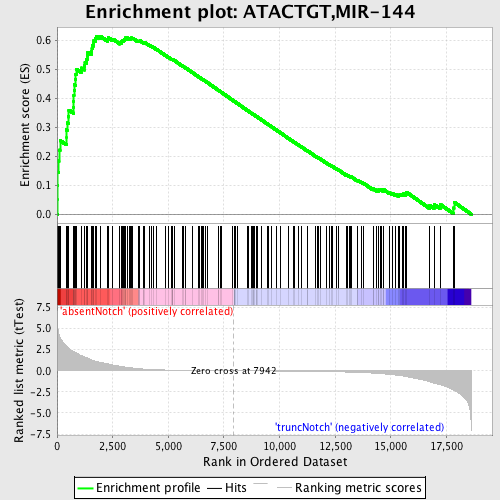
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch_repos |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#absentNotch_versus_truncNotch_repos |
| Upregulated in class | absentNotch |
| GeneSet | ATACTGT,MIR-144 |
| Enrichment Score (ES) | 0.6137406 |
| Normalized Enrichment Score (NES) | 1.8271923 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0023311956 |
| FWER p-Value | 0.012 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | APPBP2 | 5130215 | 13 | 5.776 | 0.0515 | Yes | ||
| 2 | GSPT1 | 5420050 | 18 | 5.469 | 0.1007 | Yes | ||
| 3 | MAPK6 | 6760520 | 38 | 4.847 | 0.1434 | Yes | ||
| 4 | ACBD3 | 2760170 | 59 | 4.615 | 0.1841 | Yes | ||
| 5 | SIN3A | 2190121 | 85 | 4.292 | 0.2215 | Yes | ||
| 6 | HAT1 | 3710082 | 132 | 3.945 | 0.2546 | Yes | ||
| 7 | PKNOX1 | 4210452 | 419 | 2.881 | 0.2652 | Yes | ||
| 8 | PPP1R16B | 3520300 | 423 | 2.877 | 0.2910 | Yes | ||
| 9 | MARCKS | 7040450 | 452 | 2.808 | 0.3149 | Yes | ||
| 10 | MTPN | 3940167 | 515 | 2.686 | 0.3358 | Yes | ||
| 11 | CCNE2 | 3120537 | 529 | 2.639 | 0.3589 | Yes | ||
| 12 | PABPN1 | 460519 | 735 | 2.263 | 0.3683 | Yes | ||
| 13 | IDH2 | 2680397 | 744 | 2.244 | 0.3881 | Yes | ||
| 14 | ARID1A | 2630022 1690551 4810110 | 752 | 2.236 | 0.4079 | Yes | ||
| 15 | UBE2D2 | 4590671 6510446 | 761 | 2.229 | 0.4277 | Yes | ||
| 16 | ZFX | 5900400 | 780 | 2.197 | 0.4465 | Yes | ||
| 17 | CD28 | 1400739 4210093 | 821 | 2.145 | 0.4637 | Yes | ||
| 18 | KPNA1 | 5270324 | 839 | 2.121 | 0.4820 | Yes | ||
| 19 | PHTF2 | 6590053 | 865 | 2.090 | 0.4995 | Yes | ||
| 20 | SLC23A2 | 6660722 | 1083 | 1.795 | 0.5040 | Yes | ||
| 21 | SLC7A11 | 2850138 | 1215 | 1.655 | 0.5118 | Yes | ||
| 22 | UBE2D3 | 3190452 | 1252 | 1.609 | 0.5244 | Yes | ||
| 23 | CUGBP2 | 6180121 6840139 | 1328 | 1.521 | 0.5341 | Yes | ||
| 24 | BRPF1 | 3170139 | 1352 | 1.496 | 0.5464 | Yes | ||
| 25 | EIF4G2 | 3800575 6860184 | 1379 | 1.470 | 0.5583 | Yes | ||
| 26 | NGFRAP1 | 6770195 | 1534 | 1.303 | 0.5617 | Yes | ||
| 27 | TFRC | 4050551 | 1567 | 1.269 | 0.5714 | Yes | ||
| 28 | HNRPU | 6520736 | 1587 | 1.253 | 0.5817 | Yes | ||
| 29 | EIF5 | 6380750 | 1630 | 1.216 | 0.5904 | Yes | ||
| 30 | PUM1 | 6130500 | 1654 | 1.182 | 0.5999 | Yes | ||
| 31 | ITSN2 | 780484 | 1720 | 1.139 | 0.6066 | Yes | ||
| 32 | PPP3R1 | 1190041 1570487 | 1772 | 1.092 | 0.6137 | Yes | ||
| 33 | GATA3 | 6130068 | 1948 | 0.991 | 0.6132 | No | ||
| 34 | NR3C1 | 630563 | 2279 | 0.804 | 0.6026 | No | ||
| 35 | RBM12 | 4570239 6040411 6660129 | 2303 | 0.794 | 0.6085 | No | ||
| 36 | FBXO32 | 110037 610750 | 2510 | 0.670 | 0.6034 | No | ||
| 37 | HSF2 | 1400465 | 2824 | 0.530 | 0.5913 | No | ||
| 38 | SUCLA2 | 730632 1170292 | 2890 | 0.503 | 0.5923 | No | ||
| 39 | STARD8 | 1780047 | 2893 | 0.502 | 0.5967 | No | ||
| 40 | RNF111 | 2970072 3140112 5130647 | 2948 | 0.479 | 0.5981 | No | ||
| 41 | SLC12A2 | 4610019 6040672 | 2990 | 0.468 | 0.6001 | No | ||
| 42 | ARFGEF1 | 6760494 | 3031 | 0.452 | 0.6021 | No | ||
| 43 | NEDD4 | 460736 1190280 | 3060 | 0.438 | 0.6045 | No | ||
| 44 | NFE2L2 | 2810619 3390162 | 3075 | 0.432 | 0.6076 | No | ||
| 45 | FUSIP1 | 520082 5390114 | 3143 | 0.410 | 0.6077 | No | ||
| 46 | FBXL3 | 5910142 | 3266 | 0.371 | 0.6045 | No | ||
| 47 | VKORC1L1 | 110220 1400050 | 3304 | 0.354 | 0.6056 | No | ||
| 48 | PDE4D | 2470528 6660014 | 3326 | 0.349 | 0.6077 | No | ||
| 49 | WIF1 | 7100184 | 3397 | 0.330 | 0.6069 | No | ||
| 50 | FNDC3A | 2690097 3290044 | 3635 | 0.255 | 0.5963 | No | ||
| 51 | LHX2 | 3610463 | 3677 | 0.246 | 0.5963 | No | ||
| 52 | GCNT2 | 6400162 2370609 2760309 4540465 6100280 | 3713 | 0.237 | 0.5966 | No | ||
| 53 | RIN2 | 2450184 | 3723 | 0.234 | 0.5982 | No | ||
| 54 | PFKFB2 | 3940538 7100059 | 3889 | 0.200 | 0.5911 | No | ||
| 55 | UBE4A | 6100520 | 3893 | 0.199 | 0.5927 | No | ||
| 56 | PANK1 | 670091 6220605 | 3926 | 0.191 | 0.5927 | No | ||
| 57 | ACSL4 | 510020 1240041 3190050 4540500 5570463 | 4142 | 0.155 | 0.5824 | No | ||
| 58 | SNAP91 | 3170056 5690520 | 4235 | 0.142 | 0.5787 | No | ||
| 59 | PTP4A1 | 130692 | 4343 | 0.130 | 0.5741 | No | ||
| 60 | DGCR2 | 780041 4010731 | 4455 | 0.118 | 0.5692 | No | ||
| 61 | ZCCHC2 | 460592 | 4866 | 0.085 | 0.5477 | No | ||
| 62 | ARRDC3 | 4610390 | 5005 | 0.076 | 0.5409 | No | ||
| 63 | AHCY | 4230605 | 5146 | 0.069 | 0.5340 | No | ||
| 64 | QKI | 5220093 6130044 | 5165 | 0.068 | 0.5336 | No | ||
| 65 | SMARCA1 | 4230315 | 5177 | 0.067 | 0.5336 | No | ||
| 66 | MYO1E | 3140504 | 5208 | 0.066 | 0.5326 | No | ||
| 67 | CAV2 | 5130286 5130563 | 5292 | 0.063 | 0.5287 | No | ||
| 68 | ESRRG | 4010181 | 5648 | 0.050 | 0.5099 | No | ||
| 69 | FMN2 | 450463 | 5658 | 0.050 | 0.5099 | No | ||
| 70 | LGR4 | 1660309 | 5760 | 0.046 | 0.5048 | No | ||
| 71 | RHOBTB1 | 1410010 | 5792 | 0.045 | 0.5036 | No | ||
| 72 | JPH1 | 610739 | 6081 | 0.037 | 0.4883 | No | ||
| 73 | FOSB | 1940142 | 6364 | 0.030 | 0.4733 | No | ||
| 74 | CCNK | 6980048 | 6394 | 0.029 | 0.4720 | No | ||
| 75 | SLITRK4 | 5910022 | 6507 | 0.027 | 0.4662 | No | ||
| 76 | WTAP | 4010176 | 6552 | 0.026 | 0.4640 | No | ||
| 77 | PCDH18 | 430010 | 6562 | 0.026 | 0.4638 | No | ||
| 78 | CAPZA1 | 3140390 | 6655 | 0.024 | 0.4590 | No | ||
| 79 | HNRPF | 670138 | 6739 | 0.022 | 0.4547 | No | ||
| 80 | ZIC4 | 1500082 | 7250 | 0.012 | 0.4272 | No | ||
| 81 | RGS16 | 780091 | 7267 | 0.011 | 0.4264 | No | ||
| 82 | PALM2 | 4120068 7100398 | 7350 | 0.010 | 0.4221 | No | ||
| 83 | MEIS1 | 1400575 | 7380 | 0.010 | 0.4206 | No | ||
| 84 | PPARA | 2060026 | 7862 | 0.001 | 0.3945 | No | ||
| 85 | STAG1 | 1190300 1400722 4590100 | 7962 | -0.000 | 0.3892 | No | ||
| 86 | CALCRL | 4280035 | 7966 | -0.000 | 0.3890 | No | ||
| 87 | GDF10 | 4850082 | 7989 | -0.001 | 0.3878 | No | ||
| 88 | NR2F2 | 3170609 3310577 | 8001 | -0.001 | 0.3872 | No | ||
| 89 | SP4 | 3850176 | 8094 | -0.002 | 0.3823 | No | ||
| 90 | CBX4 | 70332 | 8562 | -0.010 | 0.3571 | No | ||
| 91 | SFRP1 | 6980315 | 8599 | -0.011 | 0.3552 | No | ||
| 92 | FAM60A | 3940092 | 8756 | -0.014 | 0.3469 | No | ||
| 93 | ALDH1A3 | 2100270 | 8778 | -0.014 | 0.3459 | No | ||
| 94 | ATP2B2 | 3780397 | 8815 | -0.015 | 0.3441 | No | ||
| 95 | FBXW11 | 6450632 | 8838 | -0.015 | 0.3430 | No | ||
| 96 | MYT1 | 6770524 | 8862 | -0.015 | 0.3419 | No | ||
| 97 | PNRC1 | 360025 | 8960 | -0.017 | 0.3368 | No | ||
| 98 | PAX3 | 50551 | 8971 | -0.017 | 0.3364 | No | ||
| 99 | RARB | 430139 1410138 | 9020 | -0.018 | 0.3340 | No | ||
| 100 | PURA | 3870156 | 9192 | -0.021 | 0.3249 | No | ||
| 101 | FOXO1A | 3120670 | 9437 | -0.025 | 0.3119 | No | ||
| 102 | ANK2 | 6510546 | 9507 | -0.027 | 0.3084 | No | ||
| 103 | SS18 | 1940440 2900072 6900129 | 9629 | -0.029 | 0.3021 | No | ||
| 104 | WDFY3 | 3610041 4560333 4570273 | 9869 | -0.034 | 0.2895 | No | ||
| 105 | FMR1 | 5050075 | 10022 | -0.037 | 0.2816 | No | ||
| 106 | DMD | 1740041 3990332 | 10389 | -0.045 | 0.2622 | No | ||
| 107 | MSX1 | 2650309 | 10625 | -0.050 | 0.2499 | No | ||
| 108 | SEMA6D | 4050324 5860138 6350307 | 10682 | -0.052 | 0.2473 | No | ||
| 109 | CDH11 | 1230133 2100180 4610110 | 10844 | -0.055 | 0.2391 | No | ||
| 110 | TRIB1 | 2320435 | 10969 | -0.058 | 0.2329 | No | ||
| 111 | RFX4 | 3710100 5340215 | 11234 | -0.065 | 0.2192 | No | ||
| 112 | CPEB3 | 3940164 | 11250 | -0.066 | 0.2190 | No | ||
| 113 | NRP2 | 4070400 5860041 6650446 | 11605 | -0.077 | 0.2005 | No | ||
| 114 | LIFR | 3360068 3830102 | 11707 | -0.081 | 0.1958 | No | ||
| 115 | ALS2 | 3130546 6400070 | 11754 | -0.083 | 0.1940 | No | ||
| 116 | TBX1 | 3710133 6590121 | 11835 | -0.086 | 0.1905 | No | ||
| 117 | TTN | 2320161 4670056 6550026 | 12124 | -0.098 | 0.1758 | No | ||
| 118 | GLCCI1 | 110091 2640082 2900692 3290070 6370014 | 12223 | -0.102 | 0.1714 | No | ||
| 119 | ATXN1 | 5550156 | 12348 | -0.109 | 0.1657 | No | ||
| 120 | SMPD3 | 520164 | 12365 | -0.110 | 0.1658 | No | ||
| 121 | ST18 | 5720537 | 12571 | -0.121 | 0.1558 | No | ||
| 122 | CXCL12 | 580546 4150750 4570068 | 12660 | -0.127 | 0.1522 | No | ||
| 123 | ARID5B | 110403 | 12985 | -0.150 | 0.1360 | No | ||
| 124 | CLK1 | 1780551 2100102 2340347 2480347 | 13056 | -0.155 | 0.1336 | No | ||
| 125 | ST6GALNAC3 | 4070563 | 13148 | -0.162 | 0.1301 | No | ||
| 126 | ELL2 | 6420091 | 13189 | -0.165 | 0.1294 | No | ||
| 127 | SORCS3 | 5700309 | 13221 | -0.167 | 0.1293 | No | ||
| 128 | PIM1 | 630047 | 13494 | -0.191 | 0.1163 | No | ||
| 129 | PLAG1 | 1450142 3870139 6520039 | 13519 | -0.194 | 0.1167 | No | ||
| 130 | PAFAH1B1 | 4230333 6420121 6450066 | 13670 | -0.213 | 0.1105 | No | ||
| 131 | BACH2 | 6760131 | 13765 | -0.225 | 0.1074 | No | ||
| 132 | ETS1 | 5270278 6450717 6620465 | 14206 | -0.282 | 0.0862 | No | ||
| 133 | CDH2 | 520435 2450451 2760025 6650477 | 14221 | -0.285 | 0.0880 | No | ||
| 134 | PTHLH | 5290739 | 14375 | -0.311 | 0.0825 | No | ||
| 135 | PTGFRN | 4120524 | 14436 | -0.321 | 0.0822 | No | ||
| 136 | EDAR | 6220670 | 14459 | -0.324 | 0.0839 | No | ||
| 137 | NARG1 | 5910563 6350095 | 14518 | -0.334 | 0.0838 | No | ||
| 138 | HOXA10 | 6110397 7100458 | 14575 | -0.345 | 0.0838 | No | ||
| 139 | SEMA6A | 2690138 3060021 | 14658 | -0.359 | 0.0827 | No | ||
| 140 | PAPPA | 4230463 | 14662 | -0.361 | 0.0857 | No | ||
| 141 | CPEB2 | 4760338 | 14942 | -0.423 | 0.0745 | No | ||
| 142 | HDHD2 | 2850411 6940168 | 15084 | -0.462 | 0.0710 | No | ||
| 143 | NIN | 3450576 5080048 | 15229 | -0.510 | 0.0678 | No | ||
| 144 | CRSP2 | 1940059 | 15365 | -0.556 | 0.0655 | No | ||
| 145 | DLG5 | 450215 | 15406 | -0.570 | 0.0685 | No | ||
| 146 | CBX1 | 5080408 1980239 | 15508 | -0.604 | 0.0685 | No | ||
| 147 | PTPN9 | 3290408 | 15582 | -0.635 | 0.0703 | No | ||
| 148 | CCT2 | 1690113 | 15681 | -0.678 | 0.0711 | No | ||
| 149 | PDE7B | 5700056 | 15713 | -0.691 | 0.0757 | No | ||
| 150 | SON | 3120148 5860239 7040403 | 16751 | -1.303 | 0.0313 | No | ||
| 151 | ARID2 | 110647 5390435 | 16945 | -1.467 | 0.0341 | No | ||
| 152 | MYST3 | 5270500 | 17220 | -1.648 | 0.0341 | No | ||
| 153 | ATP5G2 | 840068 | 17831 | -2.283 | 0.0217 | No | ||
| 154 | AGRN | 6770152 | 17840 | -2.297 | 0.0420 | No |

