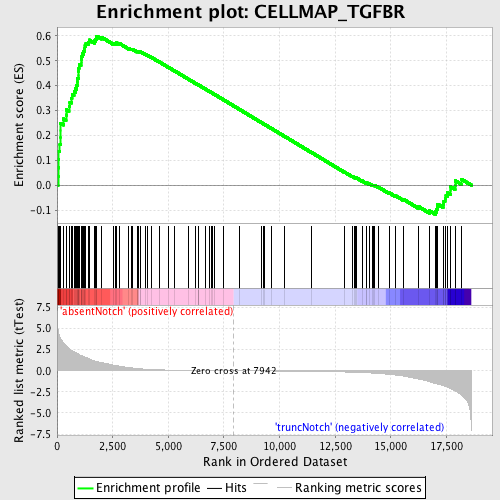
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch_repos |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#absentNotch_versus_truncNotch_repos |
| Upregulated in class | absentNotch |
| GeneSet | CELLMAP_TGFBR |
| Enrichment Score (ES) | 0.59917724 |
| Normalized Enrichment Score (NES) | 1.7064378 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.13058376 |
| FWER p-Value | 0.349 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ANAPC1 | 6350162 | 41 | 4.838 | 0.0341 | Yes | ||
| 2 | NFYB | 1850053 | 49 | 4.754 | 0.0695 | Yes | ||
| 3 | CTCF | 5340017 | 61 | 4.586 | 0.1033 | Yes | ||
| 4 | SMAD4 | 5670519 | 72 | 4.465 | 0.1363 | Yes | ||
| 5 | STRAP | 1340170 | 126 | 4.009 | 0.1636 | Yes | ||
| 6 | ANAPC4 | 4540338 | 147 | 3.854 | 0.1914 | Yes | ||
| 7 | RBL1 | 3130372 | 151 | 3.829 | 0.2200 | Yes | ||
| 8 | KPNB1 | 1690138 | 152 | 3.826 | 0.2488 | Yes | ||
| 9 | PIAS1 | 5340301 | 284 | 3.281 | 0.2663 | Yes | ||
| 10 | ZEB1 | 6840162 | 401 | 2.938 | 0.2821 | Yes | ||
| 11 | RB1 | 5900338 | 411 | 2.912 | 0.3035 | Yes | ||
| 12 | CREBBP | 5690035 7040050 | 535 | 2.625 | 0.3166 | Yes | ||
| 13 | MAPK8 | 2640195 | 574 | 2.526 | 0.3335 | Yes | ||
| 14 | NCOA1 | 3610438 | 637 | 2.418 | 0.3483 | Yes | ||
| 15 | MAP3K7 | 6040068 | 670 | 2.361 | 0.3643 | Yes | ||
| 16 | UBE2D2 | 4590671 6510446 | 761 | 2.229 | 0.3762 | Yes | ||
| 17 | NFYA | 5860368 | 831 | 2.132 | 0.3885 | Yes | ||
| 18 | UBE2E1 | 4230064 1090440 | 882 | 2.074 | 0.4014 | Yes | ||
| 19 | SMAD2 | 4200592 | 912 | 2.023 | 0.4150 | Yes | ||
| 20 | ANAPC5 | 730164 7100685 | 922 | 2.010 | 0.4296 | Yes | ||
| 21 | SNX2 | 2470068 3360364 6510064 | 964 | 1.934 | 0.4419 | Yes | ||
| 22 | PRKCD | 770592 | 966 | 1.931 | 0.4564 | Yes | ||
| 23 | SP1 | 6590017 | 975 | 1.918 | 0.4704 | Yes | ||
| 24 | TFDP1 | 1980112 | 986 | 1.906 | 0.4841 | Yes | ||
| 25 | FNTA | 70059 | 1075 | 1.808 | 0.4930 | Yes | ||
| 26 | HDAC1 | 2850670 | 1077 | 1.803 | 0.5065 | Yes | ||
| 27 | SNX4 | 1980358 | 1109 | 1.769 | 0.5181 | Yes | ||
| 28 | MAPK14 | 5290731 | 1140 | 1.736 | 0.5295 | Yes | ||
| 29 | CDC25A | 3800184 | 1196 | 1.670 | 0.5391 | Yes | ||
| 30 | CUL1 | 1990632 | 1241 | 1.620 | 0.5489 | Yes | ||
| 31 | UBE2D3 | 3190452 | 1252 | 1.609 | 0.5604 | Yes | ||
| 32 | CDC16 | 1940706 | 1295 | 1.556 | 0.5698 | Yes | ||
| 33 | DAB2 | 60309 | 1401 | 1.442 | 0.5750 | Yes | ||
| 34 | COPS5 | 1340722 5050215 | 1436 | 1.406 | 0.5837 | Yes | ||
| 35 | CCNE1 | 110064 | 1670 | 1.172 | 0.5799 | Yes | ||
| 36 | EID2 | 3130176 | 1734 | 1.128 | 0.5850 | Yes | ||
| 37 | MAP2K6 | 1230056 2940204 | 1761 | 1.102 | 0.5919 | Yes | ||
| 38 | CDK2 | 130484 2260301 4010088 5050110 | 1778 | 1.088 | 0.5992 | Yes | ||
| 39 | RBX1 | 2120010 2340047 | 1998 | 0.965 | 0.5946 | No | ||
| 40 | PRKAR2A | 2340136 | 2520 | 0.664 | 0.5714 | No | ||
| 41 | SMURF2 | 1940161 | 2624 | 0.622 | 0.5705 | No | ||
| 42 | SNW1 | 4010736 | 2659 | 0.608 | 0.5733 | No | ||
| 43 | MEF2A | 2350041 3360128 6040180 | 2811 | 0.535 | 0.5691 | No | ||
| 44 | ATF2 | 2260348 2480441 | 3226 | 0.383 | 0.5496 | No | ||
| 45 | VDR | 130156 510438 | 3329 | 0.348 | 0.5467 | No | ||
| 46 | MYC | 380541 4670170 | 3372 | 0.337 | 0.5470 | No | ||
| 47 | RBL2 | 580446 1400670 | 3592 | 0.267 | 0.5372 | No | ||
| 48 | UBE2L3 | 1690707 5900242 | 3622 | 0.258 | 0.5376 | No | ||
| 49 | ZFYVE9 | 6980288 7100017 | 3637 | 0.254 | 0.5387 | No | ||
| 50 | CDKN1A | 4050088 6400706 | 3730 | 0.231 | 0.5355 | No | ||
| 51 | CDK6 | 4920253 | 3755 | 0.226 | 0.5359 | No | ||
| 52 | CDC23 | 3190593 | 3954 | 0.186 | 0.5266 | No | ||
| 53 | MAP3K7IP1 | 2120095 5890452 6040167 | 4044 | 0.170 | 0.5230 | No | ||
| 54 | YAP1 | 4230577 | 4259 | 0.140 | 0.5125 | No | ||
| 55 | ATF3 | 1940546 | 4592 | 0.105 | 0.4954 | No | ||
| 56 | JUN | 840170 | 4993 | 0.077 | 0.4744 | No | ||
| 57 | ESR1 | 4060372 5860193 | 5289 | 0.063 | 0.4589 | No | ||
| 58 | TGFBR3 | 5290577 | 5923 | 0.042 | 0.4250 | No | ||
| 59 | UBE2D1 | 1850400 | 6210 | 0.034 | 0.4098 | No | ||
| 60 | PRKAR1B | 6130411 | 6343 | 0.030 | 0.4029 | No | ||
| 61 | SKIL | 1090364 | 6355 | 0.030 | 0.4025 | No | ||
| 62 | FOSB | 1940142 | 6364 | 0.030 | 0.4023 | No | ||
| 63 | CITED1 | 2350670 | 6682 | 0.023 | 0.3854 | No | ||
| 64 | SMAD6 | 870504 | 6850 | 0.019 | 0.3765 | No | ||
| 65 | TGFB3 | 1070041 | 6952 | 0.017 | 0.3712 | No | ||
| 66 | CCNB2 | 6510528 | 6991 | 0.017 | 0.3693 | No | ||
| 67 | HOXA9 | 610494 4730040 5130601 | 7072 | 0.016 | 0.3650 | No | ||
| 68 | XPO1 | 540707 | 7492 | 0.008 | 0.3425 | No | ||
| 69 | NUP153 | 7000452 | 8186 | -0.004 | 0.3051 | No | ||
| 70 | NFYC | 1400746 | 9183 | -0.021 | 0.2514 | No | ||
| 71 | TGFBR1 | 1400148 4280020 6550711 | 9267 | -0.022 | 0.2471 | No | ||
| 72 | SMAD7 | 430377 | 9316 | -0.023 | 0.2447 | No | ||
| 73 | CAMK2B | 2760041 | 9655 | -0.030 | 0.2266 | No | ||
| 74 | CAV1 | 870025 | 10215 | -0.041 | 0.1967 | No | ||
| 75 | ANAPC10 | 870086 1170037 2260129 | 11439 | -0.072 | 0.1312 | No | ||
| 76 | AR | 6380167 | 12908 | -0.144 | 0.0530 | No | ||
| 77 | SNX6 | 6200086 | 13259 | -0.170 | 0.0354 | No | ||
| 78 | ZEB2 | 4120070 4150215 | 13384 | -0.180 | 0.0300 | No | ||
| 79 | ENG | 4280270 | 13420 | -0.183 | 0.0295 | No | ||
| 80 | FOXO3 | 2510484 4480451 | 13441 | -0.186 | 0.0298 | No | ||
| 81 | SNX1 | 3190670 | 13705 | -0.217 | 0.0172 | No | ||
| 82 | FOS | 1850315 | 13888 | -0.238 | 0.0092 | No | ||
| 83 | RUNX2 | 1400193 4230215 | 13923 | -0.241 | 0.0092 | No | ||
| 84 | SDC2 | 1980168 | 14027 | -0.255 | 0.0055 | No | ||
| 85 | AXIN1 | 3360358 6940451 | 14175 | -0.277 | -0.0004 | No | ||
| 86 | ETS1 | 5270278 6450717 6620465 | 14206 | -0.282 | 0.0001 | No | ||
| 87 | CD44 | 3990072 4850671 5860411 6860148 7050551 | 14275 | -0.293 | -0.0013 | No | ||
| 88 | SNIP1 | 5360092 | 14451 | -0.323 | -0.0084 | No | ||
| 89 | CCND1 | 460524 770309 3120576 6980398 | 14927 | -0.419 | -0.0309 | No | ||
| 90 | TGFB1 | 1940162 | 15204 | -0.500 | -0.0420 | No | ||
| 91 | CAMK2G | 630097 | 15552 | -0.623 | -0.0561 | No | ||
| 92 | BTRC | 3170131 | 16231 | -0.978 | -0.0854 | No | ||
| 93 | FZR1 | 6220504 6660670 | 16735 | -1.294 | -0.1028 | No | ||
| 94 | STK11IP | 6980735 | 17016 | -1.506 | -0.1066 | No | ||
| 95 | TGFBR2 | 1780711 1980537 6550398 | 17052 | -1.525 | -0.0971 | No | ||
| 96 | FKBP1A | 2450368 | 17092 | -1.554 | -0.0875 | No | ||
| 97 | CAMK2D | 2370685 | 17105 | -1.563 | -0.0764 | No | ||
| 98 | SMAD3 | 6450671 | 17349 | -1.764 | -0.0763 | No | ||
| 99 | ANAPC2 | 6660438 | 17382 | -1.790 | -0.0646 | No | ||
| 100 | TP53 | 6130707 | 17437 | -1.852 | -0.0536 | No | ||
| 101 | JUNB | 4230048 | 17478 | -1.886 | -0.0416 | No | ||
| 102 | CDK4 | 540075 4540600 | 17551 | -1.953 | -0.0308 | No | ||
| 103 | PIK3R2 | 1660193 | 17684 | -2.087 | -0.0222 | No | ||
| 104 | HSPA8 | 2120315 1050132 6550746 | 17686 | -2.090 | -0.0066 | No | ||
| 105 | DAXX | 4010647 | 17887 | -2.363 | 0.0003 | No | ||
| 106 | ARRB2 | 2060441 | 17894 | -2.368 | 0.0178 | No | ||
| 107 | ANAPC7 | 5260274 | 18170 | -2.815 | 0.0241 | No |

