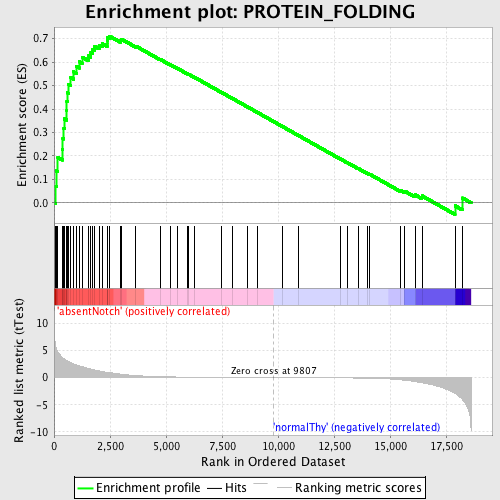
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | absentNotch |
| GeneSet | PROTEIN_FOLDING |
| Enrichment Score (ES) | 0.7110132 |
| Normalized Enrichment Score (NES) | 1.6797779 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.05712384 |
| FWER p-Value | 0.28 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RUVBL2 | 1190377 | 63 | 5.901 | 0.0717 | Yes | ||
| 2 | CCT6A | 2350368 | 104 | 5.267 | 0.1365 | Yes | ||
| 3 | DNAJC7 | 4920048 | 167 | 4.738 | 0.1935 | Yes | ||
| 4 | BAG2 | 2900324 | 371 | 3.683 | 0.2294 | Yes | ||
| 5 | ST13 | 3850369 | 390 | 3.616 | 0.2744 | Yes | ||
| 6 | FKBP4 | 870035 | 434 | 3.492 | 0.3165 | Yes | ||
| 7 | CCT3 | 7100403 | 461 | 3.376 | 0.3581 | Yes | ||
| 8 | CLN3 | 780368 | 562 | 3.127 | 0.3925 | Yes | ||
| 9 | HSPE1 | 3170438 | 569 | 3.119 | 0.4318 | Yes | ||
| 10 | STUB1 | 5860086 | 586 | 3.076 | 0.4701 | Yes | ||
| 11 | LMAN1 | 3420068 | 640 | 2.937 | 0.5046 | Yes | ||
| 12 | CCT4 | 6380161 | 744 | 2.730 | 0.5338 | Yes | ||
| 13 | TBCA | 6400095 | 873 | 2.498 | 0.5587 | Yes | ||
| 14 | ERP29 | 4050441 | 995 | 2.310 | 0.5816 | Yes | ||
| 15 | ARL2 | 5890377 | 1122 | 2.161 | 0.6023 | Yes | ||
| 16 | TBCD | 5720020 | 1252 | 1.993 | 0.6207 | Yes | ||
| 17 | GLRX2 | 940433 | 1530 | 1.648 | 0.6267 | Yes | ||
| 18 | FKBP5 | 2190048 | 1637 | 1.548 | 0.6407 | Yes | ||
| 19 | PPIH | 5910403 | 1722 | 1.471 | 0.6549 | Yes | ||
| 20 | CCT7 | 6660091 | 1807 | 1.392 | 0.6681 | Yes | ||
| 21 | PIN4 | 2370324 | 2025 | 1.198 | 0.6717 | Yes | ||
| 22 | CLPX | 6220156 | 2150 | 1.096 | 0.6789 | Yes | ||
| 23 | DNAJA3 | 1980671 6220338 | 2367 | 0.987 | 0.6798 | Yes | ||
| 24 | PFDN4 | 4010286 | 2371 | 0.984 | 0.6922 | Yes | ||
| 25 | AHSA1 | 2030520 | 2391 | 0.974 | 0.7036 | Yes | ||
| 26 | TTC1 | 5890736 | 2472 | 0.925 | 0.7110 | Yes | ||
| 27 | TXNDC4 | 450537 | 2979 | 0.629 | 0.6918 | No | ||
| 28 | PDIA6 | 2760632 | 3006 | 0.617 | 0.6982 | No | ||
| 29 | TOR1A | 670139 5890288 | 3640 | 0.371 | 0.6688 | No | ||
| 30 | BAG5 | 2230368 6420537 | 4738 | 0.160 | 0.6118 | No | ||
| 31 | APOA2 | 6510364 6860411 | 5175 | 0.121 | 0.5898 | No | ||
| 32 | SEP15 | 1780451 2060450 5890451 | 5508 | 0.101 | 0.5732 | No | ||
| 33 | APCS | 2060500 | 5935 | 0.081 | 0.5513 | No | ||
| 34 | HSP90AA1 | 4560041 5220133 2120722 | 6011 | 0.078 | 0.5483 | No | ||
| 35 | TBCE | 520692 | 6258 | 0.070 | 0.5359 | No | ||
| 36 | DNAJA1 | 1660010 2320358 6840136 | 7463 | 0.039 | 0.4716 | No | ||
| 37 | FKBP6 | 6840725 | 7954 | 0.029 | 0.4455 | No | ||
| 38 | DNAJB6 | 2810309 5570451 | 8624 | 0.018 | 0.4097 | No | ||
| 39 | FKBP9 | 1400044 | 9070 | 0.011 | 0.3859 | No | ||
| 40 | CCT6B | 3190411 | 10211 | -0.006 | 0.3246 | No | ||
| 41 | MKKS | 2760722 3390563 | 10893 | -0.017 | 0.2881 | No | ||
| 42 | HSPA4L | 5550035 | 12782 | -0.059 | 0.1872 | No | ||
| 43 | PDIA2 | 5360706 4670300 5550040 | 13099 | -0.070 | 0.1710 | No | ||
| 44 | NFYC | 1400746 | 13592 | -0.093 | 0.1457 | No | ||
| 45 | PIGK | 4540193 6350324 6860736 | 14000 | -0.120 | 0.1253 | No | ||
| 46 | ERO1L | 3060242 4780154 5220075 | 14095 | -0.128 | 0.1219 | No | ||
| 47 | UGCGL1 | 4230072 | 15470 | -0.421 | 0.0532 | No | ||
| 48 | PPIA | 2900368 4850162 | 15655 | -0.499 | 0.0496 | No | ||
| 49 | AIP | 5720300 | 16110 | -0.737 | 0.0346 | No | ||
| 50 | BAG4 | 2260070 | 16422 | -0.954 | 0.0299 | No | ||
| 51 | LRPAP1 | 5570253 | 17898 | -2.904 | -0.0126 | No | ||
| 52 | TBCC | 4810021 | 18229 | -4.023 | 0.0208 | No |

