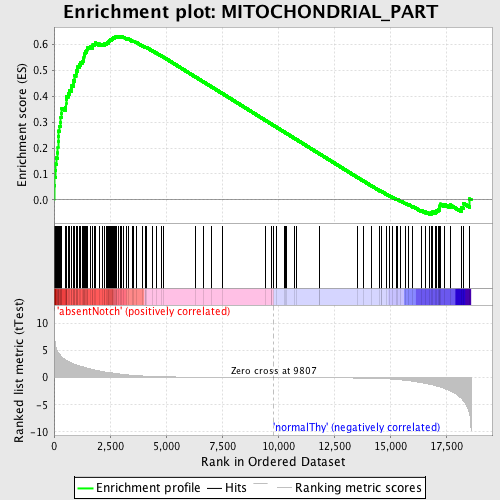
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | absentNotch |
| GeneSet | MITOCHONDRIAL_PART |
| Enrichment Score (ES) | 0.6332446 |
| Normalized Enrichment Score (NES) | 1.7109041 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0790964 |
| FWER p-Value | 0.144 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TIMM10 | 610070 1450019 | 1 | 11.709 | 0.0551 | Yes | ||
| 2 | TIMM50 | 1500139 | 33 | 6.999 | 0.0864 | Yes | ||
| 3 | TIMM13 | 2450601 | 62 | 5.924 | 0.1127 | Yes | ||
| 4 | BAX | 3830008 | 75 | 5.639 | 0.1387 | Yes | ||
| 5 | PHB2 | 1230050 | 101 | 5.289 | 0.1622 | Yes | ||
| 6 | MRPS15 | 3840280 5360504 | 164 | 4.763 | 0.1813 | Yes | ||
| 7 | ATP5G1 | 5290019 | 171 | 4.688 | 0.2030 | Yes | ||
| 8 | MRPS28 | 2230373 | 175 | 4.681 | 0.2249 | Yes | ||
| 9 | ATP5G2 | 840068 | 183 | 4.628 | 0.2463 | Yes | ||
| 10 | MRPL12 | 3120112 | 204 | 4.467 | 0.2663 | Yes | ||
| 11 | SLC25A11 | 130097 770685 | 232 | 4.319 | 0.2852 | Yes | ||
| 12 | ABCB6 | 6350138 | 292 | 4.031 | 0.3009 | Yes | ||
| 13 | BCS1L | 5360102 | 301 | 3.969 | 0.3192 | Yes | ||
| 14 | MRPL55 | 2260075 | 331 | 3.835 | 0.3357 | Yes | ||
| 15 | COX15 | 2360402 | 342 | 3.783 | 0.3530 | Yes | ||
| 16 | MRPS24 | 5340022 | 513 | 3.228 | 0.3590 | Yes | ||
| 17 | TIMM17A | 130092 | 530 | 3.187 | 0.3731 | Yes | ||
| 18 | MRPS18A | 6770433 | 534 | 3.181 | 0.3879 | Yes | ||
| 19 | VDAC2 | 7000215 | 560 | 3.129 | 0.4013 | Yes | ||
| 20 | SLC25A3 | 670112 | 631 | 2.954 | 0.4114 | Yes | ||
| 21 | SLC25A15 | 4640368 5550064 | 691 | 2.846 | 0.4216 | Yes | ||
| 22 | NDUFV1 | 5420369 | 765 | 2.694 | 0.4304 | Yes | ||
| 23 | RAB11FIP5 | 4570600 | 790 | 2.640 | 0.4415 | Yes | ||
| 24 | MRPS11 | 3130270 6660735 | 856 | 2.512 | 0.4498 | Yes | ||
| 25 | MRPS12 | 4210707 6110341 | 863 | 2.507 | 0.4613 | Yes | ||
| 26 | SUPV3L1 | 1170072 | 910 | 2.440 | 0.4703 | Yes | ||
| 27 | TOMM22 | 1240142 | 911 | 2.438 | 0.4818 | Yes | ||
| 28 | ABCF2 | 630040 3140035 6450601 | 979 | 2.325 | 0.4891 | Yes | ||
| 29 | ETFB | 2690025 | 1004 | 2.296 | 0.4986 | Yes | ||
| 30 | CYCS | 2030072 | 1055 | 2.235 | 0.5065 | Yes | ||
| 31 | MRPS10 | 2060315 4880300 6040047 | 1060 | 2.230 | 0.5167 | Yes | ||
| 32 | NDUFS2 | 4850020 6200402 | 1135 | 2.145 | 0.5228 | Yes | ||
| 33 | MRPS35 | 2370603 4070551 | 1190 | 2.073 | 0.5297 | Yes | ||
| 34 | MRPS18C | 6760044 | 1251 | 1.995 | 0.5358 | Yes | ||
| 35 | UQCRC1 | 520053 | 1300 | 1.940 | 0.5424 | Yes | ||
| 36 | SLC25A1 | 4050402 | 1330 | 1.902 | 0.5498 | Yes | ||
| 37 | ABCB7 | 4150324 | 1335 | 1.898 | 0.5585 | Yes | ||
| 38 | UQCRB | 1450047 | 1371 | 1.849 | 0.5653 | Yes | ||
| 39 | ATP5C1 | 5910601 | 1387 | 1.828 | 0.5731 | Yes | ||
| 40 | VDAC3 | 5390088 | 1459 | 1.729 | 0.5774 | Yes | ||
| 41 | ETFA | 50347 5690040 6370044 | 1469 | 1.719 | 0.5850 | Yes | ||
| 42 | NDUFA6 | 3990348 | 1504 | 1.671 | 0.5910 | Yes | ||
| 43 | CS | 5080600 | 1623 | 1.562 | 0.5920 | Yes | ||
| 44 | MRPS16 | 2510390 | 1729 | 1.465 | 0.5932 | Yes | ||
| 45 | MRPL10 | 4120484 | 1733 | 1.460 | 0.5999 | Yes | ||
| 46 | ATP5E | 4120411 | 1818 | 1.385 | 0.6019 | Yes | ||
| 47 | ACN9 | 6660180 | 1828 | 1.378 | 0.6079 | Yes | ||
| 48 | PIN4 | 2370324 | 2025 | 1.198 | 0.6029 | Yes | ||
| 49 | TIMM44 | 4670746 4730050 | 2145 | 1.099 | 0.6017 | Yes | ||
| 50 | VDAC1 | 4760059 | 2241 | 1.029 | 0.6014 | Yes | ||
| 51 | SDHD | 2570128 | 2260 | 1.023 | 0.6052 | Yes | ||
| 52 | ATP5J | 4210100 6900288 | 2343 | 0.997 | 0.6055 | Yes | ||
| 53 | DNAJA3 | 1980671 6220338 | 2367 | 0.987 | 0.6089 | Yes | ||
| 54 | NDUFA9 | 2260280 | 2432 | 0.951 | 0.6099 | Yes | ||
| 55 | ATP5D | 6550167 | 2442 | 0.944 | 0.6139 | Yes | ||
| 56 | NDUFS7 | 1740142 | 2482 | 0.914 | 0.6161 | Yes | ||
| 57 | ATP5F1 | 7000725 | 2496 | 0.905 | 0.6196 | Yes | ||
| 58 | TOMM34 | 5690102 | 2579 | 0.860 | 0.6192 | Yes | ||
| 59 | SURF1 | 5130048 | 2596 | 0.845 | 0.6224 | Yes | ||
| 60 | RAF1 | 1770600 | 2612 | 0.832 | 0.6255 | Yes | ||
| 61 | NR3C1 | 630563 | 2648 | 0.813 | 0.6274 | Yes | ||
| 62 | ATP5A1 | 1990722 | 2697 | 0.778 | 0.6285 | Yes | ||
| 63 | GRPEL1 | 3180240 | 2732 | 0.759 | 0.6302 | Yes | ||
| 64 | MRPL52 | 1170204 | 2775 | 0.732 | 0.6314 | Yes | ||
| 65 | OPA1 | 2030433 3520390 4730537 | 2804 | 0.717 | 0.6332 | Yes | ||
| 66 | TIMM23 | 60288 7040133 | 2880 | 0.680 | 0.6324 | No | ||
| 67 | BCKDK | 1940601 3990600 | 2951 | 0.649 | 0.6317 | No | ||
| 68 | NDUFS8 | 4150100 | 2987 | 0.625 | 0.6327 | No | ||
| 69 | CENTA2 | 4920451 | 3096 | 0.573 | 0.6296 | No | ||
| 70 | AIFM2 | 360537 3780520 4230138 6130082 | 3245 | 0.509 | 0.6240 | No | ||
| 71 | PMPCA | 4280132 6840408 | 3299 | 0.487 | 0.6234 | No | ||
| 72 | COX6B2 | 1340010 4120039 | 3502 | 0.416 | 0.6144 | No | ||
| 73 | BCKDHB | 780372 | 3541 | 0.405 | 0.6143 | No | ||
| 74 | MRPL51 | 540670 1580026 | 3665 | 0.363 | 0.6093 | No | ||
| 75 | TIMM8B | 1190364 | 3945 | 0.288 | 0.5956 | No | ||
| 76 | TFB2M | 3830161 4670059 4760184 | 4080 | 0.258 | 0.5896 | No | ||
| 77 | HADHB | 60064 2690670 | 4081 | 0.258 | 0.5908 | No | ||
| 78 | RHOT1 | 6760692 | 4145 | 0.246 | 0.5885 | No | ||
| 79 | NDUFAB1 | 1980091 | 4369 | 0.210 | 0.5775 | No | ||
| 80 | MAOB | 2970520 3060056 | 4557 | 0.184 | 0.5682 | No | ||
| 81 | ATP5G3 | 4230551 | 4782 | 0.156 | 0.5568 | No | ||
| 82 | DBT | 5220364 5720440 | 4877 | 0.147 | 0.5524 | No | ||
| 83 | MRPL40 | 2370215 | 6304 | 0.068 | 0.4756 | No | ||
| 84 | PITRM1 | 3450400 | 6681 | 0.056 | 0.4555 | No | ||
| 85 | MRPS22 | 5910458 | 7026 | 0.047 | 0.4372 | No | ||
| 86 | POLG2 | 5690047 | 7515 | 0.037 | 0.4109 | No | ||
| 87 | BNIP3 | 3140270 | 9417 | 0.006 | 0.3081 | No | ||
| 88 | HCCS | 670397 3170341 | 9717 | 0.001 | 0.2920 | No | ||
| 89 | PHB | 2260739 | 9723 | 0.001 | 0.2917 | No | ||
| 90 | MTX2 | 6660288 | 9791 | 0.000 | 0.2881 | No | ||
| 91 | UCP3 | 4200128 4210148 | 9928 | -0.002 | 0.2807 | No | ||
| 92 | TIMM9 | 2450484 | 10266 | -0.007 | 0.2626 | No | ||
| 93 | MRPS36 | 7050072 | 10321 | -0.008 | 0.2597 | No | ||
| 94 | MFN2 | 2260195 6100164 | 10383 | -0.009 | 0.2564 | No | ||
| 95 | GATM | 380671 | 10746 | -0.015 | 0.2369 | No | ||
| 96 | SDHA | 7000056 | 10820 | -0.016 | 0.2330 | No | ||
| 97 | HSD3B2 | 70068 770037 4120114 4230170 5690333 | 11835 | -0.034 | 0.1783 | No | ||
| 98 | NDUFS1 | 2650253 | 11853 | -0.035 | 0.1776 | No | ||
| 99 | MRPL32 | 110041 | 13555 | -0.090 | 0.0860 | No | ||
| 100 | HTRA2 | 130048 1400025 | 13831 | -0.107 | 0.0716 | No | ||
| 101 | NFS1 | 1170465 2680563 | 14165 | -0.134 | 0.0543 | No | ||
| 102 | NNT | 540253 1170471 5550092 6760397 | 14542 | -0.172 | 0.0347 | No | ||
| 103 | HSD3B1 | 3060672 | 14598 | -0.182 | 0.0326 | No | ||
| 104 | OGDH | 3840333 6350100 | 14623 | -0.187 | 0.0322 | No | ||
| 105 | RHOT2 | 1660053 2120292 | 14841 | -0.226 | 0.0215 | No | ||
| 106 | ATP5O | 7000398 | 14988 | -0.262 | 0.0149 | No | ||
| 107 | NDUFA1 | 6200411 | 15101 | -0.292 | 0.0102 | No | ||
| 108 | PPOX | 2640678 3850102 | 15264 | -0.345 | 0.0030 | No | ||
| 109 | BCKDHA | 50189 | 15333 | -0.367 | 0.0011 | No | ||
| 110 | ATP5B | 3870138 | 15483 | -0.427 | -0.0050 | No | ||
| 111 | CASQ1 | 6290035 7000707 | 15682 | -0.513 | -0.0133 | No | ||
| 112 | MPV17 | 1190133 4070427 4570577 | 15815 | -0.576 | -0.0177 | No | ||
| 113 | NDUFA2 | 6020100 | 16014 | -0.681 | -0.0252 | No | ||
| 114 | SLC25A22 | 2360050 | 16409 | -0.943 | -0.0421 | No | ||
| 115 | MRPS21 | 2650008 | 16579 | -1.046 | -0.0463 | No | ||
| 116 | BCL2 | 730132 1570736 2470138 3800044 4810037 5690068 5860504 6650164 | 16774 | -1.228 | -0.0510 | No | ||
| 117 | TIMM17B | 6040400 | 16855 | -1.310 | -0.0491 | No | ||
| 118 | UQCRH | 4210463 | 16907 | -1.358 | -0.0455 | No | ||
| 119 | MRPL23 | 4070142 | 17028 | -1.483 | -0.0450 | No | ||
| 120 | ALDH4A1 | 2450450 | 17083 | -1.551 | -0.0406 | No | ||
| 121 | NDUFA13 | 6220021 | 17142 | -1.626 | -0.0361 | No | ||
| 122 | NDUFS3 | 1690136 5340528 | 17216 | -1.726 | -0.0319 | No | ||
| 123 | ACADM | 460112 3390113 | 17218 | -1.727 | -0.0239 | No | ||
| 124 | IMMT | 870450 4280438 | 17227 | -1.732 | -0.0161 | No | ||
| 125 | FIS1 | 2760017 4070286 | 17422 | -2.022 | -0.0171 | No | ||
| 126 | NDUFS4 | 510142 1240148 4760253 | 17681 | -2.452 | -0.0195 | No | ||
| 127 | ALAS2 | 6550176 | 18175 | -3.817 | -0.0282 | No | ||
| 128 | MCL1 | 1660672 | 18272 | -4.239 | -0.0134 | No | ||
| 129 | CASP7 | 4210156 | 18553 | -6.793 | 0.0034 | No |

