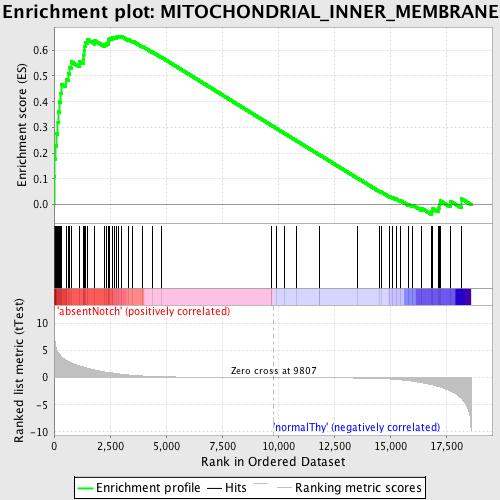
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | absentNotch |
| GeneSet | MITOCHONDRIAL_INNER_MEMBRANE |
| Enrichment Score (ES) | 0.65484655 |
| Normalized Enrichment Score (NES) | 1.5890236 |
| Nominal p-value | 0.004166667 |
| FDR q-value | 0.11696172 |
| FWER p-Value | 0.877 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TIMM10 | 610070 1450019 | 1 | 11.709 | 0.1107 | Yes | ||
| 2 | TIMM50 | 1500139 | 33 | 6.999 | 0.1752 | Yes | ||
| 3 | TIMM13 | 2450601 | 62 | 5.924 | 0.2297 | Yes | ||
| 4 | PHB2 | 1230050 | 101 | 5.289 | 0.2777 | Yes | ||
| 5 | ATP5G1 | 5290019 | 171 | 4.688 | 0.3183 | Yes | ||
| 6 | ATP5G2 | 840068 | 183 | 4.628 | 0.3615 | Yes | ||
| 7 | SLC25A11 | 130097 770685 | 232 | 4.319 | 0.3998 | Yes | ||
| 8 | BCS1L | 5360102 | 301 | 3.969 | 0.4337 | Yes | ||
| 9 | COX15 | 2360402 | 342 | 3.783 | 0.4673 | Yes | ||
| 10 | TIMM17A | 130092 | 530 | 3.187 | 0.4873 | Yes | ||
| 11 | SLC25A3 | 670112 | 631 | 2.954 | 0.5099 | Yes | ||
| 12 | SLC25A15 | 4640368 5550064 | 691 | 2.846 | 0.5336 | Yes | ||
| 13 | NDUFV1 | 5420369 | 765 | 2.694 | 0.5552 | Yes | ||
| 14 | NDUFS2 | 4850020 6200402 | 1135 | 2.145 | 0.5556 | Yes | ||
| 15 | UQCRC1 | 520053 | 1300 | 1.940 | 0.5651 | Yes | ||
| 16 | SLC25A1 | 4050402 | 1330 | 1.902 | 0.5815 | Yes | ||
| 17 | ABCB7 | 4150324 | 1335 | 1.898 | 0.5993 | Yes | ||
| 18 | UQCRB | 1450047 | 1371 | 1.849 | 0.6149 | Yes | ||
| 19 | ATP5C1 | 5910601 | 1387 | 1.828 | 0.6313 | Yes | ||
| 20 | NDUFA6 | 3990348 | 1504 | 1.671 | 0.6409 | Yes | ||
| 21 | ATP5E | 4120411 | 1818 | 1.385 | 0.6371 | Yes | ||
| 22 | SDHD | 2570128 | 2260 | 1.023 | 0.6231 | Yes | ||
| 23 | ATP5J | 4210100 6900288 | 2343 | 0.997 | 0.6281 | Yes | ||
| 24 | NDUFA9 | 2260280 | 2432 | 0.951 | 0.6323 | Yes | ||
| 25 | ATP5D | 6550167 | 2442 | 0.944 | 0.6408 | Yes | ||
| 26 | NDUFS7 | 1740142 | 2482 | 0.914 | 0.6473 | Yes | ||
| 27 | SURF1 | 5130048 | 2596 | 0.845 | 0.6492 | Yes | ||
| 28 | ATP5A1 | 1990722 | 2697 | 0.778 | 0.6512 | Yes | ||
| 29 | OPA1 | 2030433 3520390 4730537 | 2804 | 0.717 | 0.6523 | Yes | ||
| 30 | TIMM23 | 60288 7040133 | 2880 | 0.680 | 0.6547 | Yes | ||
| 31 | NDUFS8 | 4150100 | 2987 | 0.625 | 0.6548 | Yes | ||
| 32 | PMPCA | 4280132 6840408 | 3299 | 0.487 | 0.6427 | No | ||
| 33 | COX6B2 | 1340010 4120039 | 3502 | 0.416 | 0.6357 | No | ||
| 34 | TIMM8B | 1190364 | 3945 | 0.288 | 0.6146 | No | ||
| 35 | NDUFAB1 | 1980091 | 4369 | 0.210 | 0.5938 | No | ||
| 36 | ATP5G3 | 4230551 | 4782 | 0.156 | 0.5731 | No | ||
| 37 | HCCS | 670397 3170341 | 9717 | 0.001 | 0.3072 | No | ||
| 38 | PHB | 2260739 | 9723 | 0.001 | 0.3070 | No | ||
| 39 | UCP3 | 4200128 4210148 | 9928 | -0.002 | 0.2960 | No | ||
| 40 | TIMM9 | 2450484 | 10266 | -0.007 | 0.2779 | No | ||
| 41 | SDHA | 7000056 | 10820 | -0.016 | 0.2483 | No | ||
| 42 | HSD3B2 | 70068 770037 4120114 4230170 5690333 | 11835 | -0.034 | 0.1940 | No | ||
| 43 | NDUFS1 | 2650253 | 11853 | -0.035 | 0.1934 | No | ||
| 44 | MRPL32 | 110041 | 13555 | -0.090 | 0.1026 | No | ||
| 45 | NNT | 540253 1170471 5550092 6760397 | 14542 | -0.172 | 0.0510 | No | ||
| 46 | HSD3B1 | 3060672 | 14598 | -0.182 | 0.0498 | No | ||
| 47 | ATP5O | 7000398 | 14988 | -0.262 | 0.0313 | No | ||
| 48 | NDUFA1 | 6200411 | 15101 | -0.292 | 0.0281 | No | ||
| 49 | PPOX | 2640678 3850102 | 15264 | -0.345 | 0.0226 | No | ||
| 50 | ATP5B | 3870138 | 15483 | -0.427 | 0.0149 | No | ||
| 51 | MPV17 | 1190133 4070427 4570577 | 15815 | -0.576 | 0.0025 | No | ||
| 52 | NDUFA2 | 6020100 | 16014 | -0.681 | -0.0018 | No | ||
| 53 | SLC25A22 | 2360050 | 16409 | -0.943 | -0.0141 | No | ||
| 54 | TIMM17B | 6040400 | 16855 | -1.310 | -0.0257 | No | ||
| 55 | UQCRH | 4210463 | 16907 | -1.358 | -0.0156 | No | ||
| 56 | NDUFA13 | 6220021 | 17142 | -1.626 | -0.0128 | No | ||
| 57 | NDUFS3 | 1690136 5340528 | 17216 | -1.726 | -0.0004 | No | ||
| 58 | IMMT | 870450 4280438 | 17227 | -1.732 | 0.0155 | No | ||
| 59 | NDUFS4 | 510142 1240148 4760253 | 17681 | -2.452 | 0.0142 | No | ||
| 60 | ALAS2 | 6550176 | 18175 | -3.817 | 0.0238 | No |

