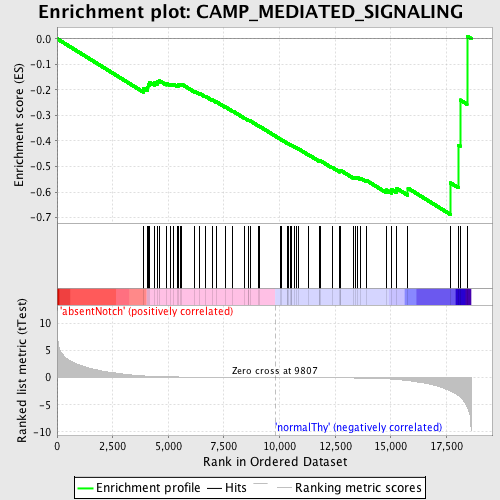
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | CAMP_MEDIATED_SIGNALING |
| Enrichment Score (ES) | -0.68790174 |
| Normalized Enrichment Score (NES) | -1.6520509 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.23106858 |
| FWER p-Value | 0.5 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ADCY7 | 6290520 7560739 | 3879 | 0.303 | -0.1937 | No | ||
| 2 | GCGR | 6620497 | 4057 | 0.263 | -0.1900 | No | ||
| 3 | ADORA2B | 6860253 | 4086 | 0.258 | -0.1785 | No | ||
| 4 | GLP1R | 6420528 | 4152 | 0.245 | -0.1696 | No | ||
| 5 | EDNRB | 4280717 6400435 | 4381 | 0.207 | -0.1715 | No | ||
| 6 | ACR | 2940017 3060050 | 4497 | 0.192 | -0.1680 | No | ||
| 7 | EDNRA | 6900133 | 4585 | 0.179 | -0.1637 | No | ||
| 8 | CCR2 | 1450519 | 4911 | 0.144 | -0.1739 | No | ||
| 9 | GLP2R | 4150577 5290273 7050095 | 5098 | 0.129 | -0.1775 | No | ||
| 10 | GRK5 | 1940348 4670053 | 5226 | 0.118 | -0.1784 | No | ||
| 11 | ADCYAP1 | 520086 | 5400 | 0.107 | -0.1823 | No | ||
| 12 | LTB4R2 | 1990746 | 5459 | 0.104 | -0.1802 | No | ||
| 13 | DRD1 | 430025 | 5530 | 0.100 | -0.1790 | No | ||
| 14 | GPR3 | 2970452 | 5590 | 0.097 | -0.1773 | No | ||
| 15 | PTGER4 | 6400670 | 6186 | 0.072 | -0.2057 | No | ||
| 16 | ADRB1 | 6860292 | 6401 | 0.065 | -0.2140 | No | ||
| 17 | CALCA | 5860167 | 6650 | 0.057 | -0.2244 | No | ||
| 18 | OPRK1 | 3780632 | 6971 | 0.048 | -0.2392 | No | ||
| 19 | DRD2 | 5890369 | 7141 | 0.045 | -0.2461 | No | ||
| 20 | CCR3 | 50427 | 7561 | 0.037 | -0.2668 | No | ||
| 21 | CAP2 | 7100315 | 7888 | 0.030 | -0.2829 | No | ||
| 22 | GALR1 | 6020452 | 8426 | 0.021 | -0.3107 | No | ||
| 23 | CHRM2 | 870750 | 8607 | 0.019 | -0.3195 | No | ||
| 24 | ADRA2A | 5340520 | 8619 | 0.018 | -0.3191 | No | ||
| 25 | MC4R | 430333 | 8673 | 0.017 | -0.3211 | No | ||
| 26 | GNAI3 | 4670739 | 9051 | 0.011 | -0.3409 | No | ||
| 27 | CALCR | 1690494 | 9113 | 0.010 | -0.3437 | No | ||
| 28 | ADORA3 | 630333 | 10031 | -0.004 | -0.3929 | No | ||
| 29 | NPY1R | 5890347 | 10103 | -0.005 | -0.3965 | No | ||
| 30 | FPR1 | 3800577 | 10334 | -0.008 | -0.4084 | No | ||
| 31 | GRM4 | 6960092 | 10371 | -0.009 | -0.4099 | No | ||
| 32 | GALR3 | 4850113 | 10407 | -0.009 | -0.4113 | No | ||
| 33 | ADRA1D | 380035 540025 | 10480 | -0.010 | -0.4147 | No | ||
| 34 | GALR2 | 6450739 | 10525 | -0.011 | -0.4165 | No | ||
| 35 | GHRHR | 3130500 4010154 | 10544 | -0.011 | -0.4169 | No | ||
| 36 | PTHLH | 5290739 | 10688 | -0.013 | -0.4239 | No | ||
| 37 | CRHR2 | 4590672 | 10741 | -0.014 | -0.4260 | No | ||
| 38 | DRD5 | 2970133 | 10847 | -0.016 | -0.4309 | No | ||
| 39 | CHRM5 | 5390333 | 11309 | -0.024 | -0.4545 | No | ||
| 40 | RGS1 | 4060347 4540181 | 11777 | -0.033 | -0.4780 | No | ||
| 41 | GRM7 | 4230110 | 11800 | -0.034 | -0.4775 | No | ||
| 42 | ADRA1B | 2100484 | 11829 | -0.034 | -0.4773 | No | ||
| 43 | ADRB3 | 6900072 | 12355 | -0.047 | -0.5032 | No | ||
| 44 | CRHR1 | 450161 3140070 5390647 | 12701 | -0.056 | -0.5189 | No | ||
| 45 | NF1 | 6980433 | 12727 | -0.057 | -0.5174 | No | ||
| 46 | AVPR2 | 360064 | 12755 | -0.058 | -0.5160 | No | ||
| 47 | GHRH | 4570575 | 13338 | -0.079 | -0.5433 | No | ||
| 48 | NPY2R | 6220612 | 13428 | -0.083 | -0.5439 | No | ||
| 49 | GRM8 | 4780082 | 13511 | -0.087 | -0.5440 | No | ||
| 50 | GRM3 | 4540242 | 13632 | -0.095 | -0.5456 | No | ||
| 51 | MCHR1 | 1780022 | 13894 | -0.113 | -0.5540 | No | ||
| 52 | GABBR1 | 2850519 2850735 | 14799 | -0.217 | -0.5918 | Yes | ||
| 53 | CORT | 360008 2340092 | 15032 | -0.273 | -0.5905 | Yes | ||
| 54 | GNAS | 630441 1850373 4050152 | 15256 | -0.341 | -0.5853 | Yes | ||
| 55 | PRKACB | 4210170 | 15771 | -0.555 | -0.5850 | Yes | ||
| 56 | NDUFS4 | 510142 1240148 4760253 | 17681 | -2.452 | -0.5642 | Yes | ||
| 57 | WASF2 | 4850592 | 18043 | -3.321 | -0.4162 | Yes | ||
| 58 | ADRB2 | 3290373 | 18117 | -3.579 | -0.2397 | Yes | ||
| 59 | CAP1 | 2650278 | 18426 | -5.284 | 0.0102 | Yes |

