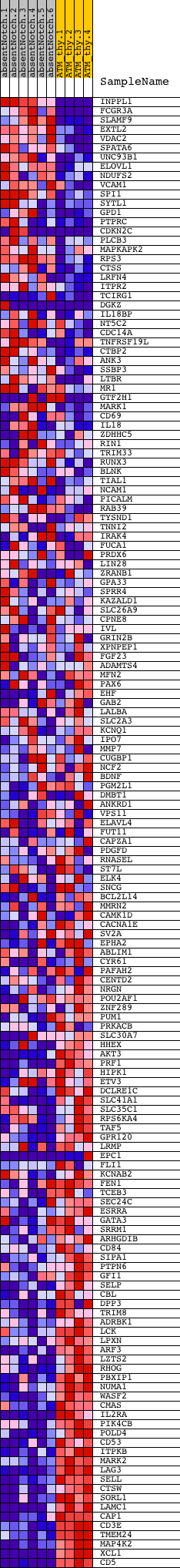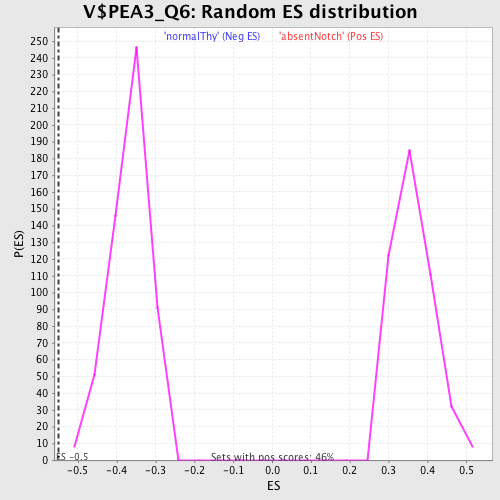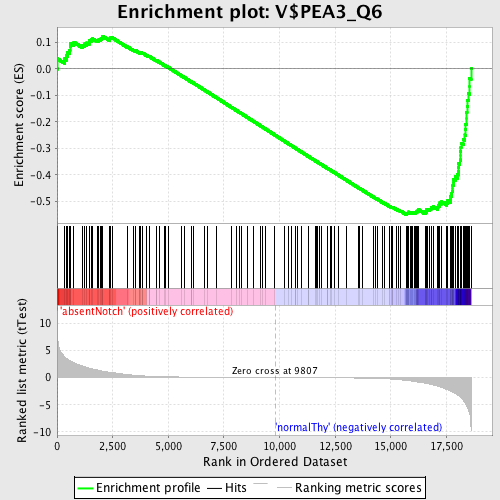
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | V$PEA3_Q6 |
| Enrichment Score (ES) | -0.5490926 |
| Normalized Enrichment Score (NES) | -1.4968026 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.14715871 |
| FWER p-Value | 0.827 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | INPPL1 | 110717 3120164 | 10 | 8.657 | 0.0385 | No | ||
| 2 | FCGR3A | 3840184 | 340 | 3.790 | 0.0378 | No | ||
| 3 | SLAMF9 | 3710300 | 424 | 3.520 | 0.0492 | No | ||
| 4 | EXTL2 | 5050433 | 467 | 3.358 | 0.0620 | No | ||
| 5 | VDAC2 | 7000215 | 560 | 3.129 | 0.0712 | No | ||
| 6 | SPATA6 | 4590373 | 581 | 3.091 | 0.0840 | No | ||
| 7 | UNC93B1 | 430152 | 600 | 3.038 | 0.0968 | No | ||
| 8 | ELOVL1 | 2450019 4760138 5340685 | 755 | 2.717 | 0.1007 | No | ||
| 9 | NDUFS2 | 4850020 6200402 | 1135 | 2.145 | 0.0898 | No | ||
| 10 | VCAM1 | 2900450 | 1226 | 2.029 | 0.0941 | No | ||
| 11 | SPI1 | 1410397 | 1318 | 1.921 | 0.0979 | No | ||
| 12 | SYTL1 | 510487 | 1438 | 1.751 | 0.0993 | No | ||
| 13 | GPD1 | 2480095 | 1444 | 1.744 | 0.1069 | No | ||
| 14 | PTPRC | 130402 5290148 | 1532 | 1.647 | 0.1096 | No | ||
| 15 | CDKN2C | 5050750 5130148 | 1569 | 1.604 | 0.1149 | No | ||
| 16 | PLCB3 | 4670402 | 1798 | 1.402 | 0.1089 | No | ||
| 17 | MAPKAPK2 | 2850500 | 1878 | 1.333 | 0.1106 | No | ||
| 18 | RPS3 | 1570017 2370170 | 1943 | 1.271 | 0.1129 | No | ||
| 19 | CTSS | 1740056 | 2004 | 1.210 | 0.1151 | No | ||
| 20 | LRFN4 | 5670368 | 2043 | 1.187 | 0.1184 | No | ||
| 21 | ITPR2 | 5360128 | 2044 | 1.185 | 0.1237 | No | ||
| 22 | TCIRG1 | 4590524 5910079 6900619 | 2359 | 0.991 | 0.1112 | No | ||
| 23 | DGKZ | 3460041 6400129 | 2376 | 0.981 | 0.1147 | No | ||
| 24 | IL18BP | 3450128 | 2397 | 0.972 | 0.1180 | No | ||
| 25 | NT5C2 | 4280735 4670528 | 2485 | 0.911 | 0.1174 | No | ||
| 26 | CDC14A | 4050132 | 3165 | 0.538 | 0.0831 | No | ||
| 27 | TNFRSF19L | 1170168 1190358 5670270 | 3441 | 0.436 | 0.0702 | No | ||
| 28 | CTBP2 | 430309 3710079 | 3520 | 0.412 | 0.0678 | No | ||
| 29 | ANK3 | 1500161 2900435 4210154 6400100 | 3523 | 0.411 | 0.0695 | No | ||
| 30 | SSBP3 | 870717 1050377 7040551 | 3708 | 0.350 | 0.0611 | No | ||
| 31 | LTBR | 2450341 | 3717 | 0.346 | 0.0623 | No | ||
| 32 | MR1 | 3850037 | 3743 | 0.338 | 0.0624 | No | ||
| 33 | GTF2H1 | 2570746 2650148 | 3831 | 0.315 | 0.0591 | No | ||
| 34 | MARK1 | 450484 | 3843 | 0.312 | 0.0600 | No | ||
| 35 | CD69 | 380167 4730088 | 4039 | 0.269 | 0.0506 | No | ||
| 36 | IL18 | 6180039 | 4139 | 0.248 | 0.0464 | No | ||
| 37 | ZDHHC5 | 1690711 | 4159 | 0.244 | 0.0464 | No | ||
| 38 | RIN1 | 510593 | 4468 | 0.196 | 0.0306 | No | ||
| 39 | TRIM33 | 580619 2230280 3990433 6200747 | 4472 | 0.196 | 0.0313 | No | ||
| 40 | RUNX3 | 3800546 | 4582 | 0.179 | 0.0263 | No | ||
| 41 | BLNK | 70288 6590092 | 4809 | 0.153 | 0.0147 | No | ||
| 42 | TIAL1 | 4150048 6510605 | 4890 | 0.146 | 0.0110 | No | ||
| 43 | NCAM1 | 3140026 | 5005 | 0.136 | 0.0055 | No | ||
| 44 | PICALM | 1940280 | 5577 | 0.097 | -0.0250 | No | ||
| 45 | RAB39 | 430524 | 5710 | 0.090 | -0.0318 | No | ||
| 46 | TYSND1 | 2940273 | 6042 | 0.077 | -0.0494 | No | ||
| 47 | TNNI2 | 5910739 | 6050 | 0.077 | -0.0494 | No | ||
| 48 | IRAK4 | 5390156 6130594 | 6133 | 0.074 | -0.0535 | No | ||
| 49 | FUCA1 | 610075 | 6610 | 0.059 | -0.0790 | No | ||
| 50 | PRDX6 | 4920397 6380601 | 6772 | 0.053 | -0.0875 | No | ||
| 51 | LIN28 | 6590672 | 7153 | 0.044 | -0.1079 | No | ||
| 52 | ZRANB1 | 510373 4540278 | 7834 | 0.031 | -0.1446 | No | ||
| 53 | GPA33 | 2650044 | 8075 | 0.027 | -0.1575 | No | ||
| 54 | SPRR4 | 5720273 | 8209 | 0.024 | -0.1646 | No | ||
| 55 | KAZALD1 | 380603 | 8288 | 0.023 | -0.1687 | No | ||
| 56 | SLC26A9 | 3440722 5340088 7510093 | 8544 | 0.020 | -0.1824 | No | ||
| 57 | CPNE8 | 7040039 | 8804 | 0.015 | -0.1964 | No | ||
| 58 | IVL | 4560500 | 9140 | 0.010 | -0.2145 | No | ||
| 59 | GRIN2B | 3800333 | 9217 | 0.009 | -0.2186 | No | ||
| 60 | XPNPEP1 | 2640750 3850315 6200242 | 9366 | 0.007 | -0.2266 | No | ||
| 61 | FGF23 | 510746 | 9792 | 0.000 | -0.2496 | No | ||
| 62 | ADAMTS4 | 430435 | 10230 | -0.007 | -0.2732 | No | ||
| 63 | MFN2 | 2260195 6100164 | 10383 | -0.009 | -0.2814 | No | ||
| 64 | PAX6 | 1190025 | 10551 | -0.011 | -0.2904 | No | ||
| 65 | EHF | 4560088 | 10721 | -0.014 | -0.2995 | No | ||
| 66 | GAB2 | 1410280 2340520 4280040 | 10728 | -0.014 | -0.2998 | No | ||
| 67 | LALBA | 6940685 | 10823 | -0.016 | -0.3048 | No | ||
| 68 | SLC2A3 | 1990377 | 10983 | -0.018 | -0.3133 | No | ||
| 69 | KCNQ1 | 1580050 6860441 | 11279 | -0.024 | -0.3292 | No | ||
| 70 | IPO7 | 2190746 | 11290 | -0.024 | -0.3296 | No | ||
| 71 | MMP7 | 1780497 | 11598 | -0.030 | -0.3461 | No | ||
| 72 | CUGBP1 | 450292 510022 7050176 7050215 | 11648 | -0.031 | -0.3486 | No | ||
| 73 | NCF2 | 540129 2370441 2650133 | 11707 | -0.032 | -0.3516 | No | ||
| 74 | BDNF | 2940128 3520368 | 11802 | -0.034 | -0.3566 | No | ||
| 75 | PGM2L1 | 2350035 | 11872 | -0.035 | -0.3602 | No | ||
| 76 | DMBT1 | 7050270 | 12154 | -0.042 | -0.3752 | No | ||
| 77 | ANKRD1 | 4850685 | 12163 | -0.042 | -0.3754 | No | ||
| 78 | VPS11 | 1090048 6040372 | 12273 | -0.045 | -0.3811 | No | ||
| 79 | ELAVL4 | 50735 3360086 5220167 | 12326 | -0.046 | -0.3838 | No | ||
| 80 | FUT11 | 6040692 | 12463 | -0.049 | -0.3909 | No | ||
| 81 | CAPZA1 | 3140390 | 12663 | -0.055 | -0.4014 | No | ||
| 82 | PDGFD | 60438 | 13006 | -0.066 | -0.4197 | No | ||
| 83 | RNASEL | 2340133 | 13552 | -0.090 | -0.4488 | No | ||
| 84 | ST7L | 840735 2360576 | 13613 | -0.094 | -0.4516 | No | ||
| 85 | ELK4 | 460494 940369 4480136 | 13711 | -0.099 | -0.4564 | No | ||
| 86 | SNCG | 3120725 | 14236 | -0.141 | -0.4842 | No | ||
| 87 | BCL2L14 | 2940279 | 14318 | -0.148 | -0.4879 | No | ||
| 88 | MMRN2 | 1740047 | 14381 | -0.154 | -0.4906 | No | ||
| 89 | CAMK1D | 1660100 2350309 | 14633 | -0.188 | -0.5033 | No | ||
| 90 | CACNA1E | 840253 | 14711 | -0.199 | -0.5066 | No | ||
| 91 | SV2A | 1450039 5570685 | 14955 | -0.252 | -0.5186 | No | ||
| 92 | EPHA2 | 5890056 | 15022 | -0.269 | -0.5210 | No | ||
| 93 | ABLIM1 | 580170 3710338 6520504 | 15053 | -0.278 | -0.5213 | No | ||
| 94 | CYR61 | 1240408 5290026 4120452 6550008 | 15080 | -0.286 | -0.5215 | No | ||
| 95 | PAFAH2 | 3140451 5720593 6510014 | 15254 | -0.341 | -0.5293 | No | ||
| 96 | CENTD2 | 60408 2510156 6100494 | 15353 | -0.373 | -0.5329 | No | ||
| 97 | NRGN | 4280433 | 15449 | -0.413 | -0.5362 | No | ||
| 98 | POU2AF1 | 5690242 | 15688 | -0.514 | -0.5468 | Yes | ||
| 99 | ZNF289 | 3610390 3870711 3940138 5340373 | 15718 | -0.530 | -0.5460 | Yes | ||
| 100 | PUM1 | 6130500 | 15727 | -0.533 | -0.5440 | Yes | ||
| 101 | PRKACB | 4210170 | 15771 | -0.555 | -0.5438 | Yes | ||
| 102 | SLC30A7 | 2230463 | 15787 | -0.562 | -0.5421 | Yes | ||
| 103 | HHEX | 2340575 | 15790 | -0.564 | -0.5396 | Yes | ||
| 104 | AKT3 | 1580270 3290278 | 15882 | -0.607 | -0.5418 | Yes | ||
| 105 | PRF1 | 6660309 | 15945 | -0.637 | -0.5423 | Yes | ||
| 106 | HIPK1 | 110193 | 15981 | -0.663 | -0.5412 | Yes | ||
| 107 | ETV3 | 5360487 | 16067 | -0.705 | -0.5427 | Yes | ||
| 108 | DCLRE1C | 1450041 2360091 3290671 3390739 | 16094 | -0.725 | -0.5408 | Yes | ||
| 109 | SLC41A1 | 1230528 | 16142 | -0.755 | -0.5399 | Yes | ||
| 110 | SLC35C1 | 4540154 | 16161 | -0.766 | -0.5375 | Yes | ||
| 111 | RPS6KA4 | 2190524 | 16206 | -0.802 | -0.5362 | Yes | ||
| 112 | TAF5 | 3450288 5890193 6860435 | 16218 | -0.813 | -0.5331 | Yes | ||
| 113 | GPR120 | 2350398 | 16239 | -0.830 | -0.5305 | Yes | ||
| 114 | LRMP | 6290193 | 16541 | -1.027 | -0.5422 | Yes | ||
| 115 | EPC1 | 1570050 5290095 6900193 | 16571 | -1.040 | -0.5390 | Yes | ||
| 116 | FLI1 | 3990142 | 16600 | -1.061 | -0.5358 | Yes | ||
| 117 | KCNAB2 | 4850315 | 16604 | -1.064 | -0.5311 | Yes | ||
| 118 | FEN1 | 1770541 | 16647 | -1.102 | -0.5284 | Yes | ||
| 119 | TCEB3 | 5890156 | 16758 | -1.211 | -0.5289 | Yes | ||
| 120 | SEC24C | 6980707 | 16808 | -1.262 | -0.5259 | Yes | ||
| 121 | ESRRA | 6550242 | 16823 | -1.271 | -0.5209 | Yes | ||
| 122 | GATA3 | 6130068 | 16926 | -1.380 | -0.5202 | Yes | ||
| 123 | SRRM1 | 6110025 | 17118 | -1.597 | -0.5234 | Yes | ||
| 124 | ARHGDIB | 4730433 | 17124 | -1.603 | -0.5164 | Yes | ||
| 125 | CD84 | 1780168 | 17198 | -1.704 | -0.5127 | Yes | ||
| 126 | SIPA1 | 5220687 | 17208 | -1.715 | -0.5054 | Yes | ||
| 127 | PTPN6 | 4230128 | 17289 | -1.812 | -0.5016 | Yes | ||
| 128 | GFI1 | 730180 | 17513 | -2.145 | -0.5040 | Yes | ||
| 129 | SELP | 6040193 | 17561 | -2.240 | -0.4964 | Yes | ||
| 130 | CBL | 6380068 | 17677 | -2.437 | -0.4917 | Yes | ||
| 131 | DPP3 | 2680725 3780670 | 17695 | -2.488 | -0.4814 | Yes | ||
| 132 | TRIM8 | 6130441 | 17722 | -2.539 | -0.4713 | Yes | ||
| 133 | ADRBK1 | 1340333 | 17749 | -2.593 | -0.4610 | Yes | ||
| 134 | LCK | 3360142 | 17786 | -2.667 | -0.4509 | Yes | ||
| 135 | LPXN | 2360142 | 17788 | -2.669 | -0.4389 | Yes | ||
| 136 | ARF3 | 3710446 | 17803 | -2.693 | -0.4276 | Yes | ||
| 137 | LZTS2 | 2260440 | 17810 | -2.702 | -0.4157 | Yes | ||
| 138 | RHOG | 6760575 | 17892 | -2.890 | -0.4070 | Yes | ||
| 139 | PBXIP1 | 70131 | 17983 | -3.134 | -0.3978 | Yes | ||
| 140 | NUMA1 | 6520576 | 18029 | -3.288 | -0.3854 | Yes | ||
| 141 | WASF2 | 4850592 | 18043 | -3.321 | -0.3711 | Yes | ||
| 142 | CMAS | 2190129 | 18053 | -3.354 | -0.3565 | Yes | ||
| 143 | IL2RA | 6620450 | 18109 | -3.560 | -0.3434 | Yes | ||
| 144 | PIK4CB | 510450 1780452 | 18127 | -3.608 | -0.3280 | Yes | ||
| 145 | POLD4 | 2320082 | 18129 | -3.623 | -0.3117 | Yes | ||
| 146 | CD53 | 5550162 | 18151 | -3.708 | -0.2961 | Yes | ||
| 147 | ITPKB | 2900195 7100039 | 18158 | -3.744 | -0.2796 | Yes | ||
| 148 | MARK2 | 7210608 | 18263 | -4.196 | -0.2663 | Yes | ||
| 149 | LAG3 | 6860112 | 18333 | -4.606 | -0.2492 | Yes | ||
| 150 | SELL | 1190500 | 18349 | -4.708 | -0.2288 | Yes | ||
| 151 | CTSW | 360368 | 18365 | -4.804 | -0.2080 | Yes | ||
| 152 | SORL1 | 630204 | 18393 | -5.016 | -0.1868 | Yes | ||
| 153 | LAMC1 | 4730025 | 18395 | -5.043 | -0.1641 | Yes | ||
| 154 | CAP1 | 2650278 | 18426 | -5.284 | -0.1419 | Yes | ||
| 155 | CD3E | 3800056 | 18448 | -5.536 | -0.1180 | Yes | ||
| 156 | TMEM24 | 5860039 | 18490 | -6.013 | -0.0931 | Yes | ||
| 157 | MAP4K2 | 4200037 | 18533 | -6.506 | -0.0661 | Yes | ||
| 158 | XCL1 | 3800504 | 18537 | -6.532 | -0.0368 | Yes | ||
| 159 | CD5 | 6590338 | 18606 | -9.083 | 0.0005 | Yes |