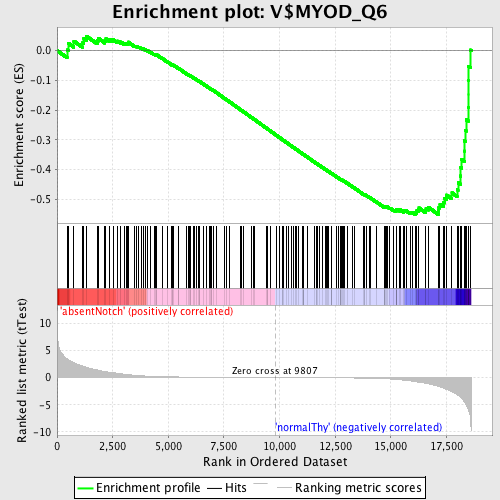
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | V$MYOD_Q6 |
| Enrichment Score (ES) | -0.55028 |
| Normalized Enrichment Score (NES) | -1.5060624 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.14686264 |
| FWER p-Value | 0.781 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CKM | 1450524 | 450 | 3.421 | 0.0021 | No | ||
| 2 | SREBF2 | 3390692 | 514 | 3.228 | 0.0236 | No | ||
| 3 | ELOVL1 | 2450019 4760138 5340685 | 755 | 2.717 | 0.0316 | No | ||
| 4 | PPP2R4 | 670280 2900097 | 1156 | 2.115 | 0.0263 | No | ||
| 5 | HMGA1 | 6580408 | 1201 | 2.059 | 0.0399 | No | ||
| 6 | SPI1 | 1410397 | 1318 | 1.921 | 0.0484 | No | ||
| 7 | WNT6 | 4570364 | 1804 | 1.397 | 0.0330 | No | ||
| 8 | CASKIN2 | 2030113 | 1841 | 1.370 | 0.0416 | No | ||
| 9 | TNFRSF21 | 6380100 | 2148 | 1.096 | 0.0335 | No | ||
| 10 | HES6 | 540411 6550504 | 2195 | 1.056 | 0.0392 | No | ||
| 11 | IGF2 | 6510020 | 2373 | 0.983 | 0.0372 | No | ||
| 12 | PRKACA | 2640731 4050048 | 2519 | 0.891 | 0.0362 | No | ||
| 13 | SCG2 | 5670438 130671 | 2707 | 0.772 | 0.0321 | No | ||
| 14 | CHCHD3 | 4200044 | 2864 | 0.686 | 0.0289 | No | ||
| 15 | ATOH7 | 5550121 | 3022 | 0.610 | 0.0251 | No | ||
| 16 | PKP4 | 3710400 5890097 | 3132 | 0.554 | 0.0235 | No | ||
| 17 | H2AFZ | 1470168 | 3180 | 0.533 | 0.0251 | No | ||
| 18 | ITPR3 | 4010632 | 3189 | 0.529 | 0.0287 | No | ||
| 19 | STK3 | 7100427 | 3498 | 0.417 | 0.0153 | No | ||
| 20 | STRN3 | 1450093 6200685 | 3563 | 0.393 | 0.0148 | No | ||
| 21 | FBXO32 | 110037 610750 | 3672 | 0.361 | 0.0118 | No | ||
| 22 | INVS | 4200632 6620528 | 3780 | 0.327 | 0.0085 | No | ||
| 23 | SYNGR1 | 5130452 6650056 | 3875 | 0.304 | 0.0058 | No | ||
| 24 | TUFT1 | 540047 2370010 | 3972 | 0.280 | 0.0027 | No | ||
| 25 | USF1 | 4280156 4610114 6370113 | 4065 | 0.261 | -0.0002 | No | ||
| 26 | ACTN3 | 3140541 6480598 | 4208 | 0.235 | -0.0061 | No | ||
| 27 | DSCAM | 1780050 2450731 2810438 | 4362 | 0.212 | -0.0128 | No | ||
| 28 | NR2E1 | 4730288 | 4420 | 0.202 | -0.0143 | No | ||
| 29 | RHOBTB1 | 1410010 | 4447 | 0.198 | -0.0142 | No | ||
| 30 | RIN1 | 510593 | 4468 | 0.196 | -0.0137 | No | ||
| 31 | MMP11 | 1980619 | 4718 | 0.163 | -0.0260 | No | ||
| 32 | NBL1 | 2480021 | 4982 | 0.138 | -0.0392 | No | ||
| 33 | LCN9 | 5050450 | 5161 | 0.122 | -0.0479 | No | ||
| 34 | HR | 2690095 6200300 | 5198 | 0.120 | -0.0489 | No | ||
| 35 | SEMA4C | 1980315 | 5199 | 0.120 | -0.0480 | No | ||
| 36 | CMYA1 | 6900632 | 5239 | 0.117 | -0.0492 | No | ||
| 37 | PPARGC1A | 4670040 | 5446 | 0.104 | -0.0595 | No | ||
| 38 | HDAC9 | 1990010 2260133 | 5455 | 0.104 | -0.0592 | No | ||
| 39 | STARD13 | 540722 | 5827 | 0.085 | -0.0786 | No | ||
| 40 | TTN | 2320161 4670056 6550026 | 5921 | 0.082 | -0.0830 | No | ||
| 41 | NPPC | 2320647 | 5948 | 0.081 | -0.0838 | No | ||
| 42 | GUCY1A3 | 110253 2640735 4070037 | 5975 | 0.080 | -0.0846 | No | ||
| 43 | CHRND | 840403 2260670 | 5983 | 0.079 | -0.0844 | No | ||
| 44 | HS6ST3 | 5910138 | 6151 | 0.073 | -0.0928 | No | ||
| 45 | INHA | 6100102 | 6183 | 0.072 | -0.0940 | No | ||
| 46 | MLLT6 | 3870168 | 6273 | 0.069 | -0.0983 | No | ||
| 47 | MYOZ3 | 1990138 | 6335 | 0.067 | -0.1010 | No | ||
| 48 | CDH1 | 1940736 | 6397 | 0.065 | -0.1038 | No | ||
| 49 | CER1 | 840102 | 6399 | 0.065 | -0.1034 | No | ||
| 50 | PAK3 | 4210136 | 6598 | 0.059 | -0.1137 | No | ||
| 51 | GRID2 | 2680242 | 6700 | 0.056 | -0.1187 | No | ||
| 52 | GATA2 | 6590280 | 6839 | 0.052 | -0.1258 | No | ||
| 53 | SLC12A5 | 1980692 | 6878 | 0.051 | -0.1275 | No | ||
| 54 | PLCB1 | 2570050 7100102 | 6935 | 0.049 | -0.1301 | No | ||
| 55 | TBR1 | 5130053 | 6949 | 0.049 | -0.1304 | No | ||
| 56 | EMX1 | 6130537 | 7017 | 0.047 | -0.1337 | No | ||
| 57 | LIN28 | 6590672 | 7153 | 0.044 | -0.1407 | No | ||
| 58 | PTPRJ | 4010707 | 7521 | 0.037 | -0.1603 | No | ||
| 59 | MAP3K13 | 3190017 | 7591 | 0.036 | -0.1637 | No | ||
| 60 | HSD11B2 | 5900053 | 7736 | 0.033 | -0.1713 | No | ||
| 61 | CHRNG | 5860273 | 8259 | 0.024 | -0.1994 | No | ||
| 62 | KAZALD1 | 380603 | 8288 | 0.023 | -0.2007 | No | ||
| 63 | COL4A4 | 1050541 | 8368 | 0.022 | -0.2049 | No | ||
| 64 | NHLH1 | 6100452 | 8743 | 0.016 | -0.2250 | No | ||
| 65 | FGF17 | 3130022 | 8847 | 0.014 | -0.2305 | No | ||
| 66 | MID1 | 4070114 4760458 | 8880 | 0.014 | -0.2321 | No | ||
| 67 | DCT | 1090347 3840494 | 9393 | 0.006 | -0.2598 | No | ||
| 68 | ASB14 | 5860427 | 9435 | 0.006 | -0.2620 | No | ||
| 69 | ANK2 | 6510546 | 9438 | 0.006 | -0.2620 | No | ||
| 70 | KCNE1L | 4670092 | 9611 | 0.003 | -0.2713 | No | ||
| 71 | ASB15 | 450563 | 9870 | -0.001 | -0.2853 | No | ||
| 72 | ANKRD2 | 6590427 | 9995 | -0.003 | -0.2920 | No | ||
| 73 | TAC4 | 4730070 | 10125 | -0.005 | -0.2990 | No | ||
| 74 | SYT4 | 5080193 | 10150 | -0.005 | -0.3002 | No | ||
| 75 | RND1 | 5080300 | 10181 | -0.006 | -0.3018 | No | ||
| 76 | NKX6-2 | 2450332 3120333 | 10294 | -0.008 | -0.3078 | No | ||
| 77 | GALR3 | 4850113 | 10407 | -0.009 | -0.3138 | No | ||
| 78 | ABL2 | 580021 | 10533 | -0.011 | -0.3205 | No | ||
| 79 | COL4A3 | 5910075 | 10618 | -0.012 | -0.3250 | No | ||
| 80 | FSCN2 | 1230039 | 10707 | -0.014 | -0.3296 | No | ||
| 81 | ERBB4 | 4610400 4810017 | 10737 | -0.014 | -0.3311 | No | ||
| 82 | EIF2C1 | 6900551 | 10860 | -0.016 | -0.3376 | No | ||
| 83 | CDON | 540204 | 11029 | -0.019 | -0.3465 | No | ||
| 84 | DSCAML1 | 1850403 5550079 | 11079 | -0.020 | -0.3490 | No | ||
| 85 | B3GALT2 | 430136 | 11083 | -0.020 | -0.3490 | No | ||
| 86 | RIMS2 | 670725 | 11093 | -0.020 | -0.3494 | No | ||
| 87 | CCNL2 | 6520167 | 11275 | -0.024 | -0.3590 | No | ||
| 88 | ARL4A | 6290091 | 11579 | -0.029 | -0.3752 | No | ||
| 89 | CUGBP1 | 450292 510022 7050176 7050215 | 11648 | -0.031 | -0.3786 | No | ||
| 90 | ALG6 | 4060131 | 11675 | -0.031 | -0.3798 | No | ||
| 91 | KCNQ4 | 3800438 7050400 | 11715 | -0.032 | -0.3817 | No | ||
| 92 | FOXI1 | 3710095 6620195 | 11772 | -0.033 | -0.3845 | No | ||
| 93 | RASGRF2 | 1990047 | 11910 | -0.036 | -0.3916 | No | ||
| 94 | TREX2 | 4920707 | 12076 | -0.040 | -0.4002 | No | ||
| 95 | USP15 | 610592 3520504 | 12094 | -0.040 | -0.4008 | No | ||
| 96 | TRPV3 | 540341 | 12132 | -0.042 | -0.4025 | No | ||
| 97 | SOST | 1170195 | 12191 | -0.043 | -0.4053 | No | ||
| 98 | PROX1 | 4210193 | 12322 | -0.046 | -0.4120 | No | ||
| 99 | ELAVL4 | 50735 3360086 5220167 | 12326 | -0.046 | -0.4118 | No | ||
| 100 | LRAT | 5130603 | 12339 | -0.046 | -0.4121 | No | ||
| 101 | DPF3 | 2650128 2970168 | 12579 | -0.052 | -0.4247 | No | ||
| 102 | TRIM3 | 5570156 | 12632 | -0.054 | -0.4271 | No | ||
| 103 | HTR7 | 2680176 | 12728 | -0.057 | -0.4318 | No | ||
| 104 | GGN | 1410528 4210082 5290170 | 12732 | -0.057 | -0.4315 | No | ||
| 105 | POU3F3 | 6400047 | 12758 | -0.058 | -0.4324 | No | ||
| 106 | FBXW11 | 6450632 | 12804 | -0.059 | -0.4344 | No | ||
| 107 | NEB | 580735 | 12816 | -0.060 | -0.4345 | No | ||
| 108 | NPEPPS | 2630731 | 12870 | -0.061 | -0.4369 | No | ||
| 109 | HEYL | 5860528 | 12938 | -0.064 | -0.4401 | No | ||
| 110 | SMARCA5 | 6620050 | 13032 | -0.067 | -0.4446 | No | ||
| 111 | ERBB3 | 5890372 | 13275 | -0.076 | -0.4571 | No | ||
| 112 | EFNA1 | 3840672 | 13359 | -0.080 | -0.4610 | No | ||
| 113 | ELAVL3 | 2850014 | 13765 | -0.102 | -0.4822 | No | ||
| 114 | CYP26A1 | 380022 2690600 | 13810 | -0.106 | -0.4837 | No | ||
| 115 | PHACTR3 | 3850435 5900445 | 13838 | -0.108 | -0.4844 | No | ||
| 116 | TJP1 | 6350184 | 13886 | -0.112 | -0.4860 | No | ||
| 117 | PRKAG2 | 2340100 | 14037 | -0.123 | -0.4932 | No | ||
| 118 | NOL4 | 5050446 5360519 | 14076 | -0.126 | -0.4943 | No | ||
| 119 | TAGAP | 3290433 | 14354 | -0.152 | -0.5081 | No | ||
| 120 | BLR1 | 1500300 5420017 | 14702 | -0.198 | -0.5254 | No | ||
| 121 | TGM1 | 1090500 | 14727 | -0.202 | -0.5251 | No | ||
| 122 | MDGA1 | 1170139 | 14732 | -0.203 | -0.5238 | No | ||
| 123 | ITGB4 | 1740021 3840482 | 14739 | -0.205 | -0.5225 | No | ||
| 124 | FUT8 | 1340068 2340056 | 14817 | -0.220 | -0.5250 | No | ||
| 125 | MTUS1 | 780348 4920609 | 14828 | -0.223 | -0.5238 | No | ||
| 126 | BCL9 | 7100112 | 14956 | -0.252 | -0.5288 | No | ||
| 127 | CRAT | 540020 2060364 3520148 | 15138 | -0.300 | -0.5362 | No | ||
| 128 | POFUT1 | 1570458 | 15234 | -0.332 | -0.5388 | No | ||
| 129 | NLK | 2030010 2450041 | 15237 | -0.333 | -0.5364 | No | ||
| 130 | GNAS | 630441 1850373 4050152 | 15256 | -0.341 | -0.5347 | No | ||
| 131 | ADAM11 | 1050008 3130494 | 15272 | -0.349 | -0.5328 | No | ||
| 132 | VKORC1L1 | 110220 1400050 | 15386 | -0.388 | -0.5359 | No | ||
| 133 | MAP3K7IP2 | 2340242 | 15413 | -0.395 | -0.5343 | No | ||
| 134 | SPOP | 450035 | 15589 | -0.469 | -0.5401 | No | ||
| 135 | NCKIPSD | 2940156 | 15615 | -0.479 | -0.5378 | No | ||
| 136 | CASQ1 | 6290035 7000707 | 15682 | -0.513 | -0.5374 | No | ||
| 137 | PDGFB | 3060440 6370008 | 15889 | -0.613 | -0.5438 | No | ||
| 138 | HN1 | 3360156 | 15969 | -0.656 | -0.5430 | No | ||
| 139 | OSR1 | 1500025 | 16095 | -0.727 | -0.5442 | Yes | ||
| 140 | ACADSB | 520170 | 16154 | -0.762 | -0.5414 | Yes | ||
| 141 | ZIC4 | 1500082 | 16172 | -0.777 | -0.5363 | Yes | ||
| 142 | USP2 | 1190292 1240253 4850035 | 16232 | -0.826 | -0.5332 | Yes | ||
| 143 | CHRNB1 | 1190671 | 16265 | -0.850 | -0.5283 | Yes | ||
| 144 | KCNN3 | 6520600 6420138 | 16556 | -1.032 | -0.5360 | Yes | ||
| 145 | ARHGEF2 | 3360577 | 16580 | -1.046 | -0.5292 | Yes | ||
| 146 | GFRA1 | 3610152 | 16696 | -1.150 | -0.5265 | Yes | ||
| 147 | BEX2 | 5670433 | 17135 | -1.613 | -0.5378 | Yes | ||
| 148 | SSH2 | 360056 | 17159 | -1.657 | -0.5262 | Yes | ||
| 149 | SIPA1 | 5220687 | 17208 | -1.715 | -0.5156 | Yes | ||
| 150 | CRELD1 | 2060253 | 17369 | -1.913 | -0.5094 | Yes | ||
| 151 | AP4S1 | 4730292 | 17414 | -2.001 | -0.4963 | Yes | ||
| 152 | TLK2 | 1170161 | 17509 | -2.142 | -0.4849 | Yes | ||
| 153 | ITGA6 | 3830129 | 17748 | -2.591 | -0.4777 | Yes | ||
| 154 | NRF1 | 2650195 | 17980 | -3.111 | -0.4662 | Yes | ||
| 155 | GALNT2 | 2260279 4060239 | 18023 | -3.261 | -0.4432 | Yes | ||
| 156 | POLD4 | 2320082 | 18129 | -3.623 | -0.4209 | Yes | ||
| 157 | ATP1B1 | 3130594 | 18145 | -3.686 | -0.3932 | Yes | ||
| 158 | ARID5A | 2450133 | 18188 | -3.859 | -0.3656 | Yes | ||
| 159 | NEDD4L | 6380368 | 18303 | -4.476 | -0.3372 | Yes | ||
| 160 | GMPR | 3990022 | 18309 | -4.502 | -0.3026 | Yes | ||
| 161 | SELL | 1190500 | 18349 | -4.708 | -0.2683 | Yes | ||
| 162 | PSTPIP1 | 3190156 | 18388 | -4.968 | -0.2319 | Yes | ||
| 163 | PLAGL2 | 1770148 | 18480 | -5.805 | -0.1920 | Yes | ||
| 164 | DEF6 | 840593 | 18489 | -6.000 | -0.1460 | Yes | ||
| 165 | TMEM24 | 5860039 | 18490 | -6.013 | -0.0995 | Yes | ||
| 166 | KCNH2 | 1170451 | 18497 | -6.062 | -0.0529 | Yes | ||
| 167 | TEC | 1400576 | 18580 | -7.663 | 0.0020 | Yes |

