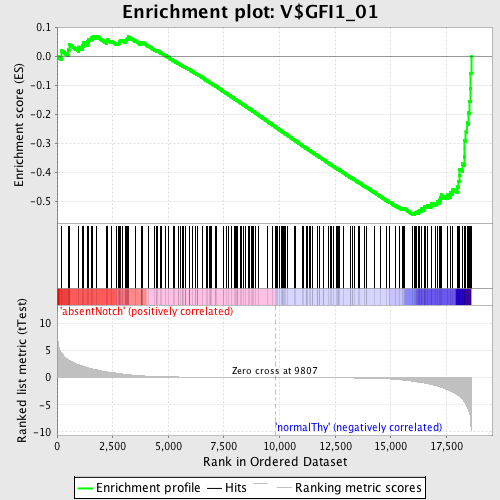
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | V$GFI1_01 |
| Enrichment Score (ES) | -0.5449416 |
| Normalized Enrichment Score (NES) | -1.5197457 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.13073343 |
| FWER p-Value | 0.714 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ATP5G2 | 840068 | 183 | 4.628 | 0.0196 | No | ||
| 2 | SREBF2 | 3390692 | 514 | 3.228 | 0.0223 | No | ||
| 3 | CSAD | 770215 | 537 | 3.174 | 0.0414 | No | ||
| 4 | ABCF2 | 630040 3140035 6450601 | 979 | 2.325 | 0.0323 | No | ||
| 5 | USP5 | 4850017 | 1138 | 2.142 | 0.0374 | No | ||
| 6 | HMGA1 | 6580408 | 1201 | 2.059 | 0.0472 | No | ||
| 7 | MYO1C | 3190398 | 1358 | 1.863 | 0.0506 | No | ||
| 8 | STMN1 | 1990717 | 1415 | 1.783 | 0.0590 | No | ||
| 9 | TMPRSS2 | 2970279 | 1526 | 1.651 | 0.0635 | No | ||
| 10 | EXOC7 | 3060037 | 1611 | 1.571 | 0.0690 | No | ||
| 11 | MCTS1 | 6130133 | 1767 | 1.433 | 0.0698 | No | ||
| 12 | MBNL1 | 2640762 7100048 | 2223 | 1.036 | 0.0517 | No | ||
| 13 | SFRS2 | 50707 380593 | 2242 | 1.028 | 0.0573 | No | ||
| 14 | SERTAD4 | 130593 | 2447 | 0.943 | 0.0522 | No | ||
| 15 | TCF4 | 520021 | 2660 | 0.803 | 0.0458 | No | ||
| 16 | MYH10 | 2640673 | 2768 | 0.735 | 0.0447 | No | ||
| 17 | TPBG | 630161 | 2803 | 0.718 | 0.0475 | No | ||
| 18 | XPO5 | 2850286 | 2815 | 0.713 | 0.0514 | No | ||
| 19 | TLK1 | 7100605 | 2845 | 0.699 | 0.0543 | No | ||
| 20 | PPARGC1B | 3290114 | 2928 | 0.659 | 0.0541 | No | ||
| 21 | CAMK2D | 2370685 | 3059 | 0.594 | 0.0508 | No | ||
| 22 | PDC | 3140400 3780184 | 3074 | 0.584 | 0.0538 | No | ||
| 23 | YWHAE | 5310435 | 3098 | 0.572 | 0.0562 | No | ||
| 24 | PPP2R2A | 2900014 5700575 | 3120 | 0.561 | 0.0586 | No | ||
| 25 | ETS1 | 5270278 6450717 6620465 | 3159 | 0.542 | 0.0600 | No | ||
| 26 | TRIM23 | 580168 6510672 | 3179 | 0.533 | 0.0624 | No | ||
| 27 | AVP | 2100113 | 3202 | 0.526 | 0.0646 | No | ||
| 28 | PRKAG1 | 6450091 | 3215 | 0.520 | 0.0672 | No | ||
| 29 | SLC4A7 | 2370056 | 3517 | 0.412 | 0.0535 | No | ||
| 30 | CDH13 | 5130368 5340114 | 3772 | 0.329 | 0.0418 | No | ||
| 31 | CNKSR1 | 730528 3390672 | 3773 | 0.329 | 0.0439 | No | ||
| 32 | CACNB2 | 1500095 7330707 | 3778 | 0.328 | 0.0458 | No | ||
| 33 | CNTN6 | 2630711 | 3804 | 0.322 | 0.0465 | No | ||
| 34 | SLC25A18 | 4540047 | 3844 | 0.311 | 0.0464 | No | ||
| 35 | RPS6KB1 | 1450427 5080110 6200563 | 3850 | 0.310 | 0.0481 | No | ||
| 36 | SOX9 | 5720681 | 4111 | 0.254 | 0.0356 | No | ||
| 37 | NR2F2 | 3170609 3310577 | 4356 | 0.213 | 0.0237 | No | ||
| 38 | LUZP1 | 940075 4280458 6180070 | 4469 | 0.196 | 0.0189 | No | ||
| 39 | PRIMA1 | 1500673 6760671 | 4470 | 0.196 | 0.0202 | No | ||
| 40 | PDE4D | 2470528 6660014 | 4523 | 0.188 | 0.0185 | No | ||
| 41 | ICAM4 | 6770053 | 4668 | 0.169 | 0.0118 | No | ||
| 42 | HOXA3 | 3610632 6020358 | 4695 | 0.166 | 0.0114 | No | ||
| 43 | MTMR4 | 4540524 | 4886 | 0.146 | 0.0021 | No | ||
| 44 | NCAM1 | 3140026 | 5005 | 0.136 | -0.0035 | No | ||
| 45 | JPH4 | 780020 | 5236 | 0.117 | -0.0152 | No | ||
| 46 | ICAM5 | 2350647 | 5283 | 0.114 | -0.0170 | No | ||
| 47 | PPARGC1A | 4670040 | 5446 | 0.104 | -0.0251 | No | ||
| 48 | ITGA10 | 6330438 | 5457 | 0.104 | -0.0250 | No | ||
| 49 | ANGPT1 | 3990368 5130288 6770035 | 5559 | 0.098 | -0.0298 | No | ||
| 50 | FOXP2 | 3520561 4150372 4760524 | 5618 | 0.095 | -0.0324 | No | ||
| 51 | TCF12 | 3610324 7000156 | 5640 | 0.094 | -0.0329 | No | ||
| 52 | GAS2 | 1340082 | 5663 | 0.093 | -0.0335 | No | ||
| 53 | OTP | 2630070 | 5748 | 0.089 | -0.0375 | No | ||
| 54 | RORA | 380164 5220215 | 5758 | 0.089 | -0.0374 | No | ||
| 55 | PEA15 | 4590685 | 5775 | 0.088 | -0.0377 | No | ||
| 56 | BAI3 | 940692 | 5928 | 0.081 | -0.0455 | No | ||
| 57 | ZFPM2 | 460072 | 5936 | 0.081 | -0.0453 | No | ||
| 58 | HOXC8 | 6220050 | 5940 | 0.081 | -0.0450 | No | ||
| 59 | MAP2K7 | 2260086 | 5956 | 0.081 | -0.0453 | No | ||
| 60 | PI15 | 3840133 | 6077 | 0.076 | -0.0513 | No | ||
| 61 | NFIA | 2760129 5860278 | 6204 | 0.071 | -0.0577 | No | ||
| 62 | HOXA9 | 610494 4730040 5130601 | 6208 | 0.071 | -0.0574 | No | ||
| 63 | GPR17 | 5890671 | 6287 | 0.069 | -0.0612 | No | ||
| 64 | HOXD11 | 2260242 | 6297 | 0.068 | -0.0612 | No | ||
| 65 | ZIC1 | 670113 | 6317 | 0.068 | -0.0618 | No | ||
| 66 | DDR2 | 7050273 | 6519 | 0.061 | -0.0724 | No | ||
| 67 | DCX | 130075 5360025 7050673 | 6732 | 0.055 | -0.0835 | No | ||
| 68 | CGN | 5890139 6370167 6400537 | 6777 | 0.053 | -0.0856 | No | ||
| 69 | ASB4 | 2100064 3060059 | 6860 | 0.051 | -0.0897 | No | ||
| 70 | ZIC3 | 380020 | 6894 | 0.051 | -0.0912 | No | ||
| 71 | HOXB8 | 3710450 | 6906 | 0.050 | -0.0914 | No | ||
| 72 | TBR1 | 5130053 | 6949 | 0.049 | -0.0934 | No | ||
| 73 | HIST1H1E | 6980706 | 7112 | 0.045 | -0.1019 | No | ||
| 74 | KLF15 | 4230164 | 7119 | 0.045 | -0.1019 | No | ||
| 75 | MEF2C | 670025 780338 | 7167 | 0.044 | -0.1042 | No | ||
| 76 | CDH6 | 4730541 | 7475 | 0.038 | -0.1206 | No | ||
| 77 | NGEF | 3190538 | 7477 | 0.038 | -0.1204 | No | ||
| 78 | PBX1 | 6660301 | 7633 | 0.035 | -0.1286 | No | ||
| 79 | GPR20 | 6620601 | 7695 | 0.034 | -0.1317 | No | ||
| 80 | BNC2 | 4810603 | 7832 | 0.031 | -0.1389 | No | ||
| 81 | SP8 | 4060576 | 7989 | 0.028 | -0.1472 | No | ||
| 82 | C1QTNF7 | 2970619 | 8028 | 0.028 | -0.1491 | No | ||
| 83 | CACNG3 | 520093 | 8070 | 0.027 | -0.1511 | No | ||
| 84 | HOXC4 | 1940193 | 8096 | 0.026 | -0.1523 | No | ||
| 85 | SH3BGRL2 | 1780239 | 8103 | 0.026 | -0.1525 | No | ||
| 86 | HOXA10 | 6110397 7100458 | 8232 | 0.024 | -0.1593 | No | ||
| 87 | PROCR | 4920687 | 8297 | 0.023 | -0.1626 | No | ||
| 88 | EBF2 | 940324 | 8355 | 0.022 | -0.1655 | No | ||
| 89 | KLF12 | 1660095 4810288 5340546 6520286 | 8472 | 0.021 | -0.1717 | No | ||
| 90 | ITSN1 | 2640577 | 8598 | 0.019 | -0.1784 | No | ||
| 91 | CLMN | 1340446 1660433 2940670 | 8637 | 0.018 | -0.1803 | No | ||
| 92 | HOXD10 | 6100039 | 8735 | 0.016 | -0.1855 | No | ||
| 93 | DSCR1L1 | 940438 | 8801 | 0.015 | -0.1889 | No | ||
| 94 | PCDH8 | 2360047 4200452 | 8827 | 0.015 | -0.1902 | No | ||
| 95 | PLCD4 | 6450040 | 8911 | 0.014 | -0.1946 | No | ||
| 96 | DEPDC2 | 5220102 | 9067 | 0.011 | -0.2029 | No | ||
| 97 | TFEC | 1170722 | 9434 | 0.006 | -0.2228 | No | ||
| 98 | CALN1 | 3610722 | 9683 | 0.002 | -0.2362 | No | ||
| 99 | TFB1M | 2190110 | 9832 | -0.000 | -0.2442 | No | ||
| 100 | PURA | 3870156 | 9881 | -0.001 | -0.2468 | No | ||
| 101 | HOXB3 | 520292 | 9890 | -0.001 | -0.2473 | No | ||
| 102 | HIST1H1T | 130632 | 9976 | -0.003 | -0.2519 | No | ||
| 103 | HOXB6 | 4210739 | 10098 | -0.005 | -0.2584 | No | ||
| 104 | DSPP | 5890403 | 10127 | -0.005 | -0.2599 | No | ||
| 105 | SOX15 | 2470603 | 10142 | -0.005 | -0.2606 | No | ||
| 106 | NRXN3 | 1400546 3360471 | 10156 | -0.006 | -0.2613 | No | ||
| 107 | GPRC5D | 4760021 | 10209 | -0.006 | -0.2641 | No | ||
| 108 | GRPR | 6020170 | 10273 | -0.007 | -0.2674 | No | ||
| 109 | PHF8 | 70722 | 10352 | -0.009 | -0.2716 | No | ||
| 110 | PTHLH | 5290739 | 10688 | -0.013 | -0.2897 | No | ||
| 111 | SGK2 | 1850070 | 10710 | -0.014 | -0.2908 | No | ||
| 112 | CDON | 540204 | 11029 | -0.019 | -0.3079 | No | ||
| 113 | BMP4 | 380113 | 11090 | -0.020 | -0.3110 | No | ||
| 114 | SLC26A3 | 380750 | 11201 | -0.022 | -0.3168 | No | ||
| 115 | GFRA3 | 6510609 | 11211 | -0.023 | -0.3172 | No | ||
| 116 | CHRM1 | 4280619 | 11235 | -0.023 | -0.3183 | No | ||
| 117 | TSGA10 | 450594 3130435 4920093 | 11347 | -0.025 | -0.3242 | No | ||
| 118 | PHYHIP | 1170377 | 11387 | -0.026 | -0.3261 | No | ||
| 119 | MSR1 | 940538 | 11474 | -0.027 | -0.3306 | No | ||
| 120 | CTLA4 | 6590537 | 11494 | -0.028 | -0.3315 | No | ||
| 121 | NKX2-8 | 50022 | 11688 | -0.031 | -0.3417 | No | ||
| 122 | RKHD3 | 6180471 | 11698 | -0.032 | -0.3420 | No | ||
| 123 | EDN1 | 1770047 | 11782 | -0.033 | -0.3463 | No | ||
| 124 | BDNF | 2940128 3520368 | 11802 | -0.034 | -0.3471 | No | ||
| 125 | POU3F1 | 3710022 | 11956 | -0.037 | -0.3552 | No | ||
| 126 | HOXA6 | 2340333 | 11979 | -0.038 | -0.3561 | No | ||
| 127 | TLR1 | 1410524 | 12176 | -0.042 | -0.3665 | No | ||
| 128 | NEXN | 1240017 5290047 | 12282 | -0.045 | -0.3719 | No | ||
| 129 | ELAVL4 | 50735 3360086 5220167 | 12326 | -0.046 | -0.3740 | No | ||
| 130 | CRYZL1 | 730022 2320044 | 12434 | -0.049 | -0.3795 | No | ||
| 131 | DPF3 | 2650128 2970168 | 12579 | -0.052 | -0.3869 | No | ||
| 132 | GREM1 | 3940180 | 12587 | -0.052 | -0.3870 | No | ||
| 133 | SPAG7 | 4560524 | 12640 | -0.054 | -0.3895 | No | ||
| 134 | MEIS1 | 1400575 | 12643 | -0.054 | -0.3892 | No | ||
| 135 | CREB5 | 2320368 | 12691 | -0.056 | -0.3914 | No | ||
| 136 | CPEB3 | 3940164 | 12871 | -0.061 | -0.4007 | No | ||
| 137 | MRGPRF | 4780746 | 13166 | -0.072 | -0.4162 | No | ||
| 138 | DSG3 | 3940014 | 13199 | -0.073 | -0.4175 | No | ||
| 139 | SOX5 | 2370576 2900167 3190128 5050528 | 13267 | -0.076 | -0.4207 | No | ||
| 140 | DNAJB4 | 380195 2760372 3940735 | 13291 | -0.077 | -0.4214 | No | ||
| 141 | GRK6 | 1500053 | 13365 | -0.080 | -0.4249 | No | ||
| 142 | SOSTDC1 | 6760017 | 13566 | -0.091 | -0.4351 | No | ||
| 143 | RARB | 430139 1410138 | 13572 | -0.091 | -0.4348 | No | ||
| 144 | PHACTR3 | 3850435 5900445 | 13838 | -0.108 | -0.4485 | No | ||
| 145 | TRDN | 1580411 | 13906 | -0.114 | -0.4514 | No | ||
| 146 | IRX3 | 4200112 | 13913 | -0.114 | -0.4510 | No | ||
| 147 | CDX2 | 3850349 | 14264 | -0.144 | -0.4691 | No | ||
| 148 | RAB6B | 5860121 | 14521 | -0.170 | -0.4819 | No | ||
| 149 | JMJD1C | 940575 2120025 | 14798 | -0.217 | -0.4955 | No | ||
| 150 | BCL9 | 7100112 | 14956 | -0.252 | -0.5024 | No | ||
| 151 | TCF1 | 5390022 | 15216 | -0.325 | -0.5144 | No | ||
| 152 | TRAF4 | 3060041 4920528 6980286 | 15390 | -0.388 | -0.5213 | No | ||
| 153 | LAMA3 | 1190131 5290239 | 15511 | -0.437 | -0.5250 | No | ||
| 154 | ELMO3 | 110372 2640019 | 15554 | -0.454 | -0.5244 | No | ||
| 155 | BPGM | 5080520 | 15609 | -0.476 | -0.5243 | No | ||
| 156 | ATF2 | 2260348 2480441 | 15990 | -0.670 | -0.5407 | Yes | ||
| 157 | PDLIM1 | 450100 | 16060 | -0.702 | -0.5399 | Yes | ||
| 158 | HIVEP3 | 3990551 | 16090 | -0.720 | -0.5369 | Yes | ||
| 159 | ZIC4 | 1500082 | 16172 | -0.777 | -0.5363 | Yes | ||
| 160 | PANK1 | 670091 6220605 | 16250 | -0.843 | -0.5351 | Yes | ||
| 161 | BRUNOL4 | 4490452 5080451 | 16298 | -0.875 | -0.5321 | Yes | ||
| 162 | RKHD1 | 6860577 | 16370 | -0.920 | -0.5301 | Yes | ||
| 163 | TOP1 | 770471 4060632 6650324 | 16388 | -0.933 | -0.5250 | Yes | ||
| 164 | HBP1 | 2510102 3130010 4210619 | 16493 | -0.998 | -0.5243 | Yes | ||
| 165 | CREM | 840156 6380438 6660041 6660168 | 16502 | -1.000 | -0.5184 | Yes | ||
| 166 | ARHGEF2 | 3360577 | 16580 | -1.046 | -0.5159 | Yes | ||
| 167 | MAP4K3 | 5390551 | 16648 | -1.104 | -0.5125 | Yes | ||
| 168 | MAP2K5 | 2030440 | 16809 | -1.262 | -0.5131 | Yes | ||
| 169 | PTEN | 3390064 | 16842 | -1.294 | -0.5066 | Yes | ||
| 170 | SOX4 | 2260091 | 16991 | -1.439 | -0.5054 | Yes | ||
| 171 | HNRPA0 | 2680048 | 17077 | -1.543 | -0.5002 | Yes | ||
| 172 | BCL9L | 6940053 | 17177 | -1.682 | -0.4948 | Yes | ||
| 173 | RHOA | 580142 5900131 5340450 | 17225 | -1.731 | -0.4863 | Yes | ||
| 174 | ADD3 | 4150739 5390600 | 17272 | -1.798 | -0.4773 | Yes | ||
| 175 | WDR23 | 2030372 | 17534 | -2.184 | -0.4775 | Yes | ||
| 176 | PREP | 6220292 | 17675 | -2.431 | -0.4696 | Yes | ||
| 177 | POLH | 2690528 | 17790 | -2.671 | -0.4587 | Yes | ||
| 178 | IRF2BP1 | 3170019 | 18010 | -3.221 | -0.4500 | Yes | ||
| 179 | HOXA7 | 5910152 | 18035 | -3.305 | -0.4302 | Yes | ||
| 180 | TCTA | 2680687 | 18070 | -3.421 | -0.4102 | Yes | ||
| 181 | GABARAP | 1450286 | 18089 | -3.487 | -0.3890 | Yes | ||
| 182 | BRD2 | 1450300 5340484 6100605 | 18221 | -3.988 | -0.3706 | Yes | ||
| 183 | GADD45A | 2900717 | 18311 | -4.505 | -0.3467 | Yes | ||
| 184 | MAP3K3 | 610685 | 18320 | -4.550 | -0.3180 | Yes | ||
| 185 | AGPAT4 | 6040079 | 18321 | -4.555 | -0.2889 | Yes | ||
| 186 | BCL6 | 940100 | 18378 | -4.903 | -0.2607 | Yes | ||
| 187 | EVA1 | 5690538 | 18424 | -5.274 | -0.2294 | Yes | ||
| 188 | HSD11B1 | 450066 5550408 | 18499 | -6.067 | -0.1947 | Yes | ||
| 189 | MAP4K2 | 4200037 | 18533 | -6.506 | -0.1549 | Yes | ||
| 190 | CCR7 | 2060687 | 18569 | -7.207 | -0.1108 | Yes | ||
| 191 | IL16 | 3850168 | 18595 | -8.481 | -0.0580 | Yes | ||
| 192 | BTG1 | 4200735 6040131 6200133 | 18611 | -9.256 | 0.0003 | Yes |

