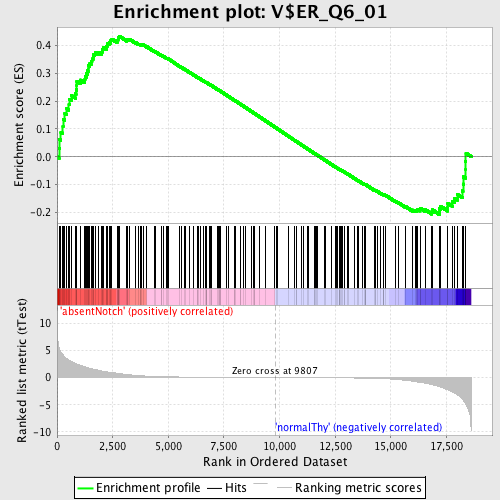
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | absentNotch |
| GeneSet | V$ER_Q6_01 |
| Enrichment Score (ES) | 0.43419102 |
| Normalized Enrichment Score (NES) | 1.2166542 |
| Nominal p-value | 0.06638116 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | EIF2B3 | 3290463 | 89 | 5.435 | 0.0297 | Yes | ||
| 2 | SLC6A9 | 2470520 | 108 | 5.216 | 0.0618 | Yes | ||
| 3 | ATP5G1 | 5290019 | 171 | 4.688 | 0.0882 | Yes | ||
| 4 | HSPG2 | 2510687 6220750 | 262 | 4.167 | 0.1098 | Yes | ||
| 5 | PCBP4 | 6350722 | 270 | 4.138 | 0.1357 | Yes | ||
| 6 | ALDOA | 6290672 | 348 | 3.761 | 0.1554 | Yes | ||
| 7 | FKBP4 | 870035 | 434 | 3.492 | 0.1729 | Yes | ||
| 8 | SDC1 | 3440471 6350408 | 533 | 3.182 | 0.1878 | Yes | ||
| 9 | RRBP1 | 2260477 5050440 | 555 | 3.137 | 0.2066 | Yes | ||
| 10 | PHF5A | 2690519 | 652 | 2.899 | 0.2198 | Yes | ||
| 11 | NR1D1 | 2360471 770746 6590204 | 825 | 2.570 | 0.2268 | Yes | ||
| 12 | PPP1R16A | 5080239 | 852 | 2.518 | 0.2413 | Yes | ||
| 13 | POLE3 | 5270019 | 882 | 2.480 | 0.2555 | Yes | ||
| 14 | ACO2 | 4230600 | 888 | 2.467 | 0.2709 | Yes | ||
| 15 | GREB1 | 1230707 4560056 | 1051 | 2.237 | 0.2763 | Yes | ||
| 16 | GTPBP1 | 2230541 4560528 | 1249 | 1.997 | 0.2783 | Yes | ||
| 17 | TOMM40 | 6110017 | 1256 | 1.984 | 0.2906 | Yes | ||
| 18 | SLC25A1 | 4050402 | 1330 | 1.902 | 0.2987 | Yes | ||
| 19 | B3GNT6 | 1340736 | 1357 | 1.869 | 0.3091 | Yes | ||
| 20 | MT2A | 6860286 | 1412 | 1.785 | 0.3175 | Yes | ||
| 21 | STMN1 | 1990717 | 1415 | 1.783 | 0.3287 | Yes | ||
| 22 | DGCR8 | 3450632 | 1473 | 1.717 | 0.3366 | Yes | ||
| 23 | NFIL3 | 4070377 | 1531 | 1.647 | 0.3439 | Yes | ||
| 24 | CDKN2C | 5050750 5130148 | 1569 | 1.604 | 0.3521 | Yes | ||
| 25 | ZFPM1 | 6760093 | 1617 | 1.566 | 0.3595 | Yes | ||
| 26 | TCF7 | 3800736 5390181 | 1628 | 1.557 | 0.3688 | Yes | ||
| 27 | RHOB | 1500309 | 1703 | 1.496 | 0.3743 | Yes | ||
| 28 | CKB | 4560632 | 1843 | 1.370 | 0.3755 | Yes | ||
| 29 | SRP68 | 4480731 | 2002 | 1.212 | 0.3746 | Yes | ||
| 30 | RNMT | 1770500 | 2022 | 1.199 | 0.3812 | Yes | ||
| 31 | LRFN4 | 5670368 | 2043 | 1.187 | 0.3876 | Yes | ||
| 32 | TRPS1 | 870100 | 2081 | 1.158 | 0.3930 | Yes | ||
| 33 | GNB1L | 3840368 5270215 | 2218 | 1.038 | 0.3922 | Yes | ||
| 34 | NDRG2 | 450403 | 2230 | 1.033 | 0.3981 | Yes | ||
| 35 | PSMF1 | 5340056 | 2249 | 1.027 | 0.4037 | Yes | ||
| 36 | SLC16A6 | 1690156 | 2278 | 1.015 | 0.4086 | Yes | ||
| 37 | SYNJ2BP | 4150672 6770706 | 2352 | 0.995 | 0.4110 | Yes | ||
| 38 | RTN4 | 50725 630647 870097 870184 | 2392 | 0.973 | 0.4150 | Yes | ||
| 39 | HIP2 | 2810095 3990369 4120301 | 2416 | 0.961 | 0.4199 | Yes | ||
| 40 | TBX6 | 4730139 6840647 | 2460 | 0.935 | 0.4235 | Yes | ||
| 41 | ATP5A1 | 1990722 | 2697 | 0.778 | 0.4156 | Yes | ||
| 42 | PDHA1 | 5550397 | 2709 | 0.771 | 0.4199 | Yes | ||
| 43 | PDE6D | 3130537 3390014 | 2737 | 0.755 | 0.4232 | Yes | ||
| 44 | BNIP3L | 1940347 | 2743 | 0.751 | 0.4277 | Yes | ||
| 45 | AAMP | 2640059 | 2777 | 0.731 | 0.4306 | Yes | ||
| 46 | RBMS1 | 6400014 | 2796 | 0.721 | 0.4342 | Yes | ||
| 47 | PKP4 | 3710400 5890097 | 3132 | 0.554 | 0.4195 | No | ||
| 48 | CDC14A | 4050132 | 3165 | 0.538 | 0.4212 | No | ||
| 49 | NR4A2 | 60273 | 3183 | 0.532 | 0.4237 | No | ||
| 50 | ZFP36L2 | 510180 | 3253 | 0.506 | 0.4231 | No | ||
| 51 | SLC4A7 | 2370056 | 3517 | 0.412 | 0.4115 | No | ||
| 52 | RAP2C | 1690132 | 3675 | 0.360 | 0.4053 | No | ||
| 53 | PDZK1 | 2320128 | 3738 | 0.340 | 0.4041 | No | ||
| 54 | SLC25A5 | 1980672 | 3761 | 0.331 | 0.4050 | No | ||
| 55 | APBA2BP | 5700341 | 3811 | 0.321 | 0.4043 | No | ||
| 56 | BRMS1 | 6860280 | 3877 | 0.304 | 0.4027 | No | ||
| 57 | MRVI1 | 4810338 4850601 5900441 | 4035 | 0.269 | 0.3959 | No | ||
| 58 | SPOCK2 | 6650369 | 4374 | 0.209 | 0.3789 | No | ||
| 59 | TLX2 | 1090128 2900451 | 4431 | 0.201 | 0.3772 | No | ||
| 60 | LCP1 | 4280397 | 4691 | 0.166 | 0.3642 | No | ||
| 61 | HOXA3 | 3610632 6020358 | 4695 | 0.166 | 0.3651 | No | ||
| 62 | NOG | 2320609 | 4770 | 0.157 | 0.3621 | No | ||
| 63 | GCM1 | 6510687 | 4912 | 0.144 | 0.3553 | No | ||
| 64 | ERG | 50154 1770739 | 4972 | 0.138 | 0.3530 | No | ||
| 65 | MCC | 620750 | 5001 | 0.136 | 0.3523 | No | ||
| 66 | FGF9 | 1050195 | 5498 | 0.101 | 0.3261 | No | ||
| 67 | GPR3 | 2970452 | 5590 | 0.097 | 0.3218 | No | ||
| 68 | TINAG | 1570609 6510215 | 5708 | 0.090 | 0.3160 | No | ||
| 69 | OTP | 2630070 | 5748 | 0.089 | 0.3144 | No | ||
| 70 | MAP2K7 | 2260086 | 5956 | 0.081 | 0.3037 | No | ||
| 71 | KIRREL3 | 2630195 | 6140 | 0.074 | 0.2943 | No | ||
| 72 | CNTN4 | 1780300 5570577 6370019 | 6305 | 0.068 | 0.2858 | No | ||
| 73 | ZIC1 | 670113 | 6317 | 0.068 | 0.2856 | No | ||
| 74 | STARD6 | 6940541 | 6367 | 0.066 | 0.2834 | No | ||
| 75 | ESRRB | 3870082 | 6427 | 0.064 | 0.2806 | No | ||
| 76 | TLX3 | 510619 | 6591 | 0.059 | 0.2721 | No | ||
| 77 | TSGA14 | 6660465 | 6687 | 0.056 | 0.2673 | No | ||
| 78 | GRID2 | 2680242 | 6700 | 0.056 | 0.2670 | No | ||
| 79 | PDE1A | 1050458 2570563 | 6728 | 0.055 | 0.2659 | No | ||
| 80 | DCX | 130075 5360025 7050673 | 6732 | 0.055 | 0.2661 | No | ||
| 81 | BHLHB5 | 6510520 | 6867 | 0.051 | 0.2592 | No | ||
| 82 | PFTK1 | 1780181 | 6873 | 0.051 | 0.2592 | No | ||
| 83 | UBE2R2 | 1230504 | 6948 | 0.049 | 0.2555 | No | ||
| 84 | TBR1 | 5130053 | 6949 | 0.049 | 0.2558 | No | ||
| 85 | PTHR1 | 2640711 3360647 | 7230 | 0.043 | 0.2409 | No | ||
| 86 | FIGN | 7000601 | 7255 | 0.043 | 0.2399 | No | ||
| 87 | NKX2-2 | 4150731 | 7312 | 0.042 | 0.2371 | No | ||
| 88 | EGFL9 | 1410091 | 7315 | 0.042 | 0.2373 | No | ||
| 89 | DNMT3A | 4050068 4810717 | 7333 | 0.041 | 0.2366 | No | ||
| 90 | GFAP | 2060092 | 7354 | 0.041 | 0.2358 | No | ||
| 91 | WNT3 | 2360685 | 7618 | 0.035 | 0.2217 | No | ||
| 92 | GNAI1 | 4560390 | 7724 | 0.033 | 0.2162 | No | ||
| 93 | FBXL14 | 4670315 | 7964 | 0.029 | 0.2035 | No | ||
| 94 | SRPK2 | 6380341 | 7985 | 0.028 | 0.2026 | No | ||
| 95 | ONECUT2 | 1340008 | 7998 | 0.028 | 0.2021 | No | ||
| 96 | HOXA10 | 6110397 7100458 | 8232 | 0.024 | 0.1896 | No | ||
| 97 | WNT11 | 1230278 | 8240 | 0.024 | 0.1894 | No | ||
| 98 | CALD1 | 1770129 1940397 | 8361 | 0.022 | 0.1830 | No | ||
| 99 | PGF | 4810593 | 8475 | 0.021 | 0.1770 | No | ||
| 100 | SLC10A2 | 110537 | 8720 | 0.017 | 0.1639 | No | ||
| 101 | PCDH8 | 2360047 4200452 | 8827 | 0.015 | 0.1582 | No | ||
| 102 | TBC1D15 | 870673 | 8853 | 0.014 | 0.1570 | No | ||
| 103 | MID1 | 4070114 4760458 | 8880 | 0.014 | 0.1556 | No | ||
| 104 | APEX2 | 3780044 | 9074 | 0.011 | 0.1452 | No | ||
| 105 | ATP6V0A4 | 360100 | 9352 | 0.007 | 0.1303 | No | ||
| 106 | PAK1IP1 | 2230438 | 9764 | 0.001 | 0.1080 | No | ||
| 107 | PURA | 3870156 | 9881 | -0.001 | 0.1017 | No | ||
| 108 | HOXB3 | 520292 | 9890 | -0.001 | 0.1013 | No | ||
| 109 | RIMS4 | 6110500 | 10394 | -0.009 | 0.0740 | No | ||
| 110 | PXN | 3290048 6400132 | 10672 | -0.013 | 0.0591 | No | ||
| 111 | ERBB4 | 4610400 4810017 | 10737 | -0.014 | 0.0557 | No | ||
| 112 | FSHB | 7100592 | 10777 | -0.015 | 0.0537 | No | ||
| 113 | HIPK2 | 6350647 | 10984 | -0.018 | 0.0426 | No | ||
| 114 | B3GALT2 | 430136 | 11083 | -0.020 | 0.0375 | No | ||
| 115 | OTX2 | 1190471 | 11239 | -0.023 | 0.0292 | No | ||
| 116 | LIFR | 3360068 3830102 | 11318 | -0.024 | 0.0251 | No | ||
| 117 | PPARA | 2060026 | 11553 | -0.029 | 0.0126 | No | ||
| 118 | SPRED1 | 6940706 | 11559 | -0.029 | 0.0125 | No | ||
| 119 | S100A5 | 4120592 | 11610 | -0.030 | 0.0100 | No | ||
| 120 | PAM | 5290528 | 11665 | -0.031 | 0.0073 | No | ||
| 121 | MITF | 380056 | 11690 | -0.031 | 0.0062 | No | ||
| 122 | PRDM1 | 3170347 3520301 | 12003 | -0.038 | -0.0105 | No | ||
| 123 | TREX2 | 4920707 | 12076 | -0.040 | -0.0142 | No | ||
| 124 | HOXB9 | 2640687 | 12334 | -0.046 | -0.0278 | No | ||
| 125 | CHRDL1 | 1580037 6380204 | 12513 | -0.050 | -0.0371 | No | ||
| 126 | SHH | 5570400 | 12546 | -0.051 | -0.0386 | No | ||
| 127 | SCNN1A | 4730019 | 12595 | -0.053 | -0.0408 | No | ||
| 128 | GNRHR | 130438 | 12678 | -0.055 | -0.0449 | No | ||
| 129 | CREB5 | 2320368 | 12691 | -0.056 | -0.0452 | No | ||
| 130 | IL23R | 3990014 | 12710 | -0.056 | -0.0458 | No | ||
| 131 | POU3F3 | 6400047 | 12758 | -0.058 | -0.0480 | No | ||
| 132 | COVA1 | 5340504 5360594 | 12793 | -0.059 | -0.0495 | No | ||
| 133 | GLRA2 | 7000019 | 12826 | -0.060 | -0.0508 | No | ||
| 134 | ISL1 | 4810239 | 12915 | -0.063 | -0.0552 | No | ||
| 135 | LRRN3 | 1450133 | 12923 | -0.063 | -0.0552 | No | ||
| 136 | CDKL5 | 2450070 | 13048 | -0.067 | -0.0615 | No | ||
| 137 | ERN1 | 2360403 | 13106 | -0.070 | -0.0641 | No | ||
| 138 | RBBP9 | 2480139 | 13356 | -0.080 | -0.0771 | No | ||
| 139 | GRM8 | 4780082 | 13511 | -0.087 | -0.0849 | No | ||
| 140 | GAS7 | 2120358 6450494 | 13563 | -0.091 | -0.0871 | No | ||
| 141 | SP6 | 60484 510452 2690333 | 13741 | -0.101 | -0.0961 | No | ||
| 142 | JARID2 | 6290538 | 13824 | -0.107 | -0.0998 | No | ||
| 143 | LRP5 | 2100397 3170484 | 13839 | -0.108 | -0.0999 | No | ||
| 144 | BTBD3 | 1980041 2510577 3190053 4200746 | 13848 | -0.109 | -0.0997 | No | ||
| 145 | LRRN1 | 3290154 | 13857 | -0.110 | -0.0994 | No | ||
| 146 | RAB11FIP1 | 2470333 | 14246 | -0.142 | -0.1195 | No | ||
| 147 | CLDN12 | 6350341 | 14306 | -0.148 | -0.1218 | No | ||
| 148 | FBXW7 | 4210338 7050280 | 14316 | -0.148 | -0.1214 | No | ||
| 149 | GNA13 | 4590102 | 14413 | -0.158 | -0.1256 | No | ||
| 150 | NNT | 540253 1170471 5550092 6760397 | 14542 | -0.172 | -0.1314 | No | ||
| 151 | DDR1 | 2060044 5220180 | 14663 | -0.192 | -0.1367 | No | ||
| 152 | TPM3 | 670600 3170296 5670167 6650471 | 14667 | -0.192 | -0.1357 | No | ||
| 153 | COPZ2 | 1780010 2360020 4070373 | 14690 | -0.196 | -0.1356 | No | ||
| 154 | AOC2 | 520348 3120008 | 14760 | -0.209 | -0.1380 | No | ||
| 155 | HOXB7 | 4540706 4610358 | 15205 | -0.323 | -0.1600 | No | ||
| 156 | EIF4E | 1580403 70133 6380215 | 15331 | -0.366 | -0.1645 | No | ||
| 157 | MNT | 4920575 | 15674 | -0.510 | -0.1798 | No | ||
| 158 | GABARAPL2 | 610204 3360047 | 15967 | -0.654 | -0.1915 | No | ||
| 159 | CAMKK2 | 5270047 | 16092 | -0.723 | -0.1936 | No | ||
| 160 | ZIC4 | 1500082 | 16172 | -0.777 | -0.1930 | No | ||
| 161 | NDUFB3 | 4610687 | 16211 | -0.809 | -0.1899 | No | ||
| 162 | DNAJA2 | 2630253 6860358 | 16312 | -0.885 | -0.1897 | No | ||
| 163 | DTNB | 2850100 | 16325 | -0.895 | -0.1847 | No | ||
| 164 | KCNN3 | 6520600 6420138 | 16556 | -1.032 | -0.1906 | No | ||
| 165 | FCHSD2 | 5720092 | 16847 | -1.299 | -0.1981 | No | ||
| 166 | SLC9A1 | 2650093 | 16874 | -1.327 | -0.1911 | No | ||
| 167 | CHD2 | 4070039 | 17182 | -1.689 | -0.1970 | No | ||
| 168 | FGF13 | 630575 1570440 5360121 | 17192 | -1.699 | -0.1867 | No | ||
| 169 | IMMT | 870450 4280438 | 17227 | -1.732 | -0.1776 | No | ||
| 170 | MAP2K6 | 1230056 2940204 | 17557 | -2.234 | -0.1812 | No | ||
| 171 | MAGED1 | 6650332 | 17564 | -2.250 | -0.1673 | No | ||
| 172 | PHF12 | 870400 3130687 | 17759 | -2.611 | -0.1612 | No | ||
| 173 | UHRF2 | 60050 840280 1340746 5080091 | 17853 | -2.807 | -0.1484 | No | ||
| 174 | EIF2AK3 | 4280161 | 17993 | -3.174 | -0.1358 | No | ||
| 175 | COX7A2L | 1500142 | 18217 | -3.972 | -0.1227 | No | ||
| 176 | CD37 | 770273 | 18246 | -4.132 | -0.0980 | No | ||
| 177 | SATB1 | 5670154 | 18264 | -4.200 | -0.0722 | No | ||
| 178 | MAP1LC3A | 840519 5570605 | 18351 | -4.713 | -0.0470 | No | ||
| 179 | CIC | 6370161 | 18357 | -4.746 | -0.0171 | No | ||
| 180 | BCL6 | 940100 | 18378 | -4.903 | 0.0129 | No |

