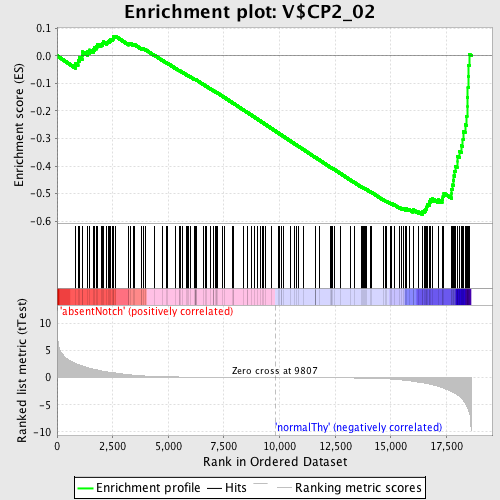
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | V$CP2_02 |
| Enrichment Score (ES) | -0.57528347 |
| Normalized Enrichment Score (NES) | -1.5527755 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.13565983 |
| FWER p-Value | 0.558 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NR1D1 | 2360471 770746 6590204 | 825 | 2.570 | -0.0277 | No | ||
| 2 | DDIT4 | 2570408 | 940 | 2.388 | -0.0182 | No | ||
| 3 | KLHDC3 | 3830717 | 992 | 2.311 | -0.0057 | No | ||
| 4 | PPP1R16B | 3520300 | 1125 | 2.158 | 0.0014 | No | ||
| 5 | MRPL14 | 2370168 | 1150 | 2.120 | 0.0141 | No | ||
| 6 | KCTD15 | 2810735 | 1372 | 1.845 | 0.0143 | No | ||
| 7 | NISCH | 4850301 | 1461 | 1.728 | 0.0210 | No | ||
| 8 | CS | 5080600 | 1623 | 1.562 | 0.0225 | No | ||
| 9 | PHPT1 | 7050537 | 1684 | 1.514 | 0.0293 | No | ||
| 10 | GRB7 | 2100471 | 1774 | 1.423 | 0.0339 | No | ||
| 11 | RYR1 | 2510114 3130408 3710086 | 1814 | 1.387 | 0.0409 | No | ||
| 12 | H3F3B | 1410300 | 1974 | 1.240 | 0.0405 | No | ||
| 13 | HTATIP | 5360739 | 2042 | 1.188 | 0.0447 | No | ||
| 14 | RASSF1 | 2760364 4850725 | 2065 | 1.165 | 0.0512 | No | ||
| 15 | MORF4L2 | 6450133 | 2235 | 1.032 | 0.0488 | No | ||
| 16 | ALDH3B1 | 4210010 6940403 | 2287 | 1.013 | 0.0528 | No | ||
| 17 | TGFBR2 | 1780711 1980537 6550398 | 2353 | 0.994 | 0.0558 | No | ||
| 18 | LHX2 | 3610463 | 2408 | 0.964 | 0.0592 | No | ||
| 19 | CAMK2G | 630097 | 2508 | 0.898 | 0.0598 | No | ||
| 20 | GMPPB | 4610538 | 2517 | 0.894 | 0.0653 | No | ||
| 21 | PRKACA | 2640731 4050048 | 2519 | 0.891 | 0.0711 | No | ||
| 22 | FKBP2 | 2320025 | 2618 | 0.830 | 0.0713 | No | ||
| 23 | PRKAG1 | 6450091 | 3215 | 0.520 | 0.0424 | No | ||
| 24 | NR2F1 | 4120528 360402 | 3230 | 0.514 | 0.0450 | No | ||
| 25 | RHBDF1 | 380538 | 3294 | 0.490 | 0.0449 | No | ||
| 26 | SHB | 4920494 | 3449 | 0.433 | 0.0394 | No | ||
| 27 | RBBP6 | 2320129 | 3493 | 0.418 | 0.0398 | No | ||
| 28 | CNTN6 | 2630711 | 3804 | 0.322 | 0.0251 | No | ||
| 29 | EPN3 | 2350592 | 3808 | 0.321 | 0.0271 | No | ||
| 30 | PPP2R5C | 3310121 6650672 | 3861 | 0.307 | 0.0263 | No | ||
| 31 | TUFT1 | 540047 2370010 | 3972 | 0.280 | 0.0222 | No | ||
| 32 | DSCAM | 1780050 2450731 2810438 | 4362 | 0.212 | 0.0025 | No | ||
| 33 | MMP11 | 1980619 | 4718 | 0.163 | -0.0156 | No | ||
| 34 | GPC3 | 6180735 | 4898 | 0.145 | -0.0244 | No | ||
| 35 | ETV1 | 70735 2940603 5080463 | 4979 | 0.138 | -0.0278 | No | ||
| 36 | RTN1 | 4150452 4850176 | 5323 | 0.112 | -0.0456 | No | ||
| 37 | FGF9 | 1050195 | 5498 | 0.101 | -0.0544 | No | ||
| 38 | STAC2 | 7100022 | 5515 | 0.100 | -0.0546 | No | ||
| 39 | SLC25A28 | 6590600 | 5561 | 0.098 | -0.0564 | No | ||
| 40 | FOXP2 | 3520561 4150372 4760524 | 5618 | 0.095 | -0.0588 | No | ||
| 41 | STARD13 | 540722 | 5827 | 0.085 | -0.0695 | No | ||
| 42 | COX8C | 4670154 | 5872 | 0.084 | -0.0713 | No | ||
| 43 | DHH | 2120433 | 5908 | 0.082 | -0.0727 | No | ||
| 44 | TBX2 | 1990563 | 5974 | 0.080 | -0.0757 | No | ||
| 45 | TNNC1 | 1990575 | 6153 | 0.073 | -0.0849 | No | ||
| 46 | FEV | 840369 5860139 | 6207 | 0.071 | -0.0873 | No | ||
| 47 | GPRC5C | 1580372 | 6209 | 0.071 | -0.0868 | No | ||
| 48 | NRIP2 | 5390288 | 6210 | 0.071 | -0.0864 | No | ||
| 49 | MAPRE3 | 7050504 | 6225 | 0.071 | -0.0867 | No | ||
| 50 | NFATC4 | 2470735 | 6245 | 0.070 | -0.0872 | No | ||
| 51 | RNF121 | 110079 1580279 | 6262 | 0.069 | -0.0876 | No | ||
| 52 | TLX3 | 510619 | 6591 | 0.059 | -0.1050 | No | ||
| 53 | EFNA3 | 6370390 | 6654 | 0.057 | -0.1080 | No | ||
| 54 | DCX | 130075 5360025 7050673 | 6732 | 0.055 | -0.1118 | No | ||
| 55 | PFTK1 | 1780181 | 6873 | 0.051 | -0.1191 | No | ||
| 56 | SRMS | 5360402 | 7010 | 0.048 | -0.1261 | No | ||
| 57 | KCNE3 | 5550195 | 7021 | 0.047 | -0.1264 | No | ||
| 58 | HPCAL1 | 1980129 2350348 | 7049 | 0.047 | -0.1275 | No | ||
| 59 | GSH1 | 2760242 | 7099 | 0.046 | -0.1299 | No | ||
| 60 | CITED1 | 2350670 | 7172 | 0.044 | -0.1335 | No | ||
| 61 | KIF1C | 2480484 | 7185 | 0.044 | -0.1338 | No | ||
| 62 | GNB3 | 1230167 | 7190 | 0.044 | -0.1338 | No | ||
| 63 | LYN | 6040600 | 7419 | 0.039 | -0.1459 | No | ||
| 64 | HSPA1L | 4010538 | 7537 | 0.037 | -0.1519 | No | ||
| 65 | PODN | 3710068 | 7874 | 0.030 | -0.1700 | No | ||
| 66 | MID2 | 4120309 | 7922 | 0.030 | -0.1723 | No | ||
| 67 | FOXL2 | 5670537 | 7941 | 0.029 | -0.1731 | No | ||
| 68 | CALD1 | 1770129 1940397 | 8361 | 0.022 | -0.1956 | No | ||
| 69 | CREB1 | 1500717 2230358 3610600 6550601 | 8536 | 0.020 | -0.2049 | No | ||
| 70 | SLC26A9 | 3440722 5340088 7510093 | 8544 | 0.020 | -0.2052 | No | ||
| 71 | UPK2 | 60176 | 8717 | 0.017 | -0.2144 | No | ||
| 72 | BRUNOL6 | 6840373 | 8863 | 0.014 | -0.2222 | No | ||
| 73 | SLC2A9 | 3290301 | 9024 | 0.012 | -0.2308 | No | ||
| 74 | AQP6 | 510736 1570484 | 9133 | 0.010 | -0.2365 | No | ||
| 75 | FRMD3 | 3440731 | 9226 | 0.009 | -0.2415 | No | ||
| 76 | TBC1D5 | 5360619 6130603 | 9296 | 0.008 | -0.2452 | No | ||
| 77 | XPNPEP1 | 2640750 3850315 6200242 | 9366 | 0.007 | -0.2489 | No | ||
| 78 | ODC1 | 5670168 | 9621 | 0.003 | -0.2626 | No | ||
| 79 | PCDHB4 | 670184 | 9934 | -0.002 | -0.2795 | No | ||
| 80 | ARHGAP5 | 2510619 3360035 | 9991 | -0.003 | -0.2825 | No | ||
| 81 | ADRA1A | 1230446 2260390 | 10081 | -0.004 | -0.2873 | No | ||
| 82 | HOXB6 | 4210739 | 10098 | -0.005 | -0.2881 | No | ||
| 83 | NRXN3 | 1400546 3360471 | 10156 | -0.006 | -0.2912 | No | ||
| 84 | ARX | 6900504 | 10475 | -0.010 | -0.3083 | No | ||
| 85 | PTHLH | 5290739 | 10688 | -0.013 | -0.3197 | No | ||
| 86 | RAB10 | 2360008 5890020 | 10753 | -0.015 | -0.3231 | No | ||
| 87 | PAK7 | 3830164 | 10835 | -0.016 | -0.3274 | No | ||
| 88 | BMP4 | 380113 | 11090 | -0.020 | -0.3410 | No | ||
| 89 | DSC2 | 840484 | 11605 | -0.030 | -0.3687 | No | ||
| 90 | NNAT | 4920053 5130253 | 11606 | -0.030 | -0.3685 | No | ||
| 91 | C1QTNF5 | 1690402 | 11617 | -0.030 | -0.3688 | No | ||
| 92 | FOXA1 | 3710609 | 11627 | -0.030 | -0.3691 | No | ||
| 93 | BDNF | 2940128 3520368 | 11802 | -0.034 | -0.3783 | No | ||
| 94 | HIF1A | 5670605 | 12295 | -0.045 | -0.4047 | No | ||
| 95 | PITX2 | 870537 2690139 | 12327 | -0.046 | -0.4060 | No | ||
| 96 | P518 | 3610435 | 12336 | -0.046 | -0.4062 | No | ||
| 97 | PAPPA | 4230463 | 12362 | -0.047 | -0.4072 | No | ||
| 98 | CAPN6 | 1740168 | 12469 | -0.049 | -0.4126 | No | ||
| 99 | TRERF1 | 6370017 6940138 | 12715 | -0.057 | -0.4255 | No | ||
| 100 | PLXNA3 | 7050164 | 13182 | -0.073 | -0.4503 | No | ||
| 101 | MOGAT2 | 60139 | 13371 | -0.080 | -0.4600 | No | ||
| 102 | WNT10B | 510050 | 13670 | -0.097 | -0.4755 | No | ||
| 103 | TXNL5 | 1190609 1400164 | 13733 | -0.101 | -0.4782 | No | ||
| 104 | GNAQ | 430670 4210131 5900736 | 13784 | -0.104 | -0.4802 | No | ||
| 105 | RPL41 | 6940112 | 13829 | -0.107 | -0.4819 | No | ||
| 106 | PAX2 | 6040270 7000133 | 13874 | -0.111 | -0.4835 | No | ||
| 107 | IRX3 | 4200112 | 13913 | -0.114 | -0.4848 | No | ||
| 108 | NOL4 | 5050446 5360519 | 14076 | -0.126 | -0.4928 | No | ||
| 109 | KIF5A | 510435 | 14128 | -0.131 | -0.4947 | No | ||
| 110 | DDR1 | 2060044 5220180 | 14663 | -0.192 | -0.5223 | No | ||
| 111 | LTBP1 | 780035 2100746 2760402 3780551 6550184 6860193 | 14776 | -0.213 | -0.5270 | No | ||
| 112 | LDB2 | 5670441 6110170 | 14821 | -0.222 | -0.5279 | No | ||
| 113 | MDK | 3840020 | 14975 | -0.258 | -0.5345 | No | ||
| 114 | NTRK3 | 4050687 4070167 4610026 6100484 | 15042 | -0.276 | -0.5362 | No | ||
| 115 | BTBD14B | 3520601 4280162 | 15153 | -0.306 | -0.5402 | No | ||
| 116 | TRAF4 | 3060041 4920528 6980286 | 15390 | -0.388 | -0.5504 | No | ||
| 117 | ZNF385 | 450176 4810358 | 15490 | -0.429 | -0.5529 | No | ||
| 118 | CPNE1 | 3440131 | 15561 | -0.457 | -0.5537 | No | ||
| 119 | MNT | 4920575 | 15674 | -0.510 | -0.5564 | No | ||
| 120 | KIFAP3 | 3800524 4590082 6940075 | 15686 | -0.514 | -0.5536 | No | ||
| 121 | FLOT2 | 50358 4210068 5080397 | 15823 | -0.578 | -0.5572 | No | ||
| 122 | DDAH2 | 4480551 | 15997 | -0.675 | -0.5621 | No | ||
| 123 | NEK8 | 5700180 | 16008 | -0.678 | -0.5582 | No | ||
| 124 | SCAMP5 | 5550368 6290021 | 16255 | -0.845 | -0.5659 | No | ||
| 125 | BACH1 | 290195 | 16429 | -0.961 | -0.5689 | Yes | ||
| 126 | YWHAG | 3780341 | 16437 | -0.964 | -0.5630 | Yes | ||
| 127 | UBE1 | 4150139 7050463 | 16518 | -1.011 | -0.5606 | Yes | ||
| 128 | EPC1 | 1570050 5290095 6900193 | 16571 | -1.040 | -0.5566 | Yes | ||
| 129 | FLI1 | 3990142 | 16600 | -1.061 | -0.5511 | Yes | ||
| 130 | CRK | 1230162 4780128 | 16625 | -1.085 | -0.5452 | Yes | ||
| 131 | MAPK3 | 580161 4780035 | 16632 | -1.091 | -0.5383 | Yes | ||
| 132 | CLDN4 | 4920739 | 16741 | -1.189 | -0.5363 | Yes | ||
| 133 | CHD4 | 5420059 6130338 6380717 | 16744 | -1.194 | -0.5286 | Yes | ||
| 134 | TLN1 | 6590411 | 16778 | -1.235 | -0.5222 | Yes | ||
| 135 | SLC9A1 | 2650093 | 16874 | -1.327 | -0.5186 | Yes | ||
| 136 | ATP10D | 4850139 | 17139 | -1.619 | -0.5222 | Yes | ||
| 137 | GSK3B | 5360348 | 17303 | -1.823 | -0.5190 | Yes | ||
| 138 | SLC25A14 | 4760180 | 17344 | -1.873 | -0.5088 | Yes | ||
| 139 | RHBDL1 | 1500736 | 17386 | -1.956 | -0.4981 | Yes | ||
| 140 | TRIM8 | 6130441 | 17722 | -2.539 | -0.4995 | Yes | ||
| 141 | MAST2 | 6200348 | 17727 | -2.546 | -0.4829 | Yes | ||
| 142 | TRPV2 | 7100348 | 17781 | -2.656 | -0.4683 | Yes | ||
| 143 | LZTS2 | 2260440 | 17810 | -2.702 | -0.4520 | Yes | ||
| 144 | HCFC1 | 2260008 | 17838 | -2.780 | -0.4351 | Yes | ||
| 145 | GLTP | 3440603 | 17866 | -2.854 | -0.4177 | Yes | ||
| 146 | PRKCH | 5720079 | 17901 | -2.912 | -0.4003 | Yes | ||
| 147 | NRF1 | 2650195 | 17980 | -3.111 | -0.3840 | Yes | ||
| 148 | DUSP6 | 5910286 7100070 | 17982 | -3.129 | -0.3634 | Yes | ||
| 149 | GABARAP | 1450286 | 18089 | -3.487 | -0.3461 | Yes | ||
| 150 | JUNB | 4230048 | 18168 | -3.777 | -0.3254 | Yes | ||
| 151 | NGFRAP1 | 6770195 | 18242 | -4.099 | -0.3023 | Yes | ||
| 152 | BCL11B | 2680673 | 18265 | -4.208 | -0.2758 | Yes | ||
| 153 | PTCRA | 6940142 | 18341 | -4.648 | -0.2491 | Yes | ||
| 154 | FBS1 | 2570520 | 18411 | -5.173 | -0.2187 | Yes | ||
| 155 | CAP1 | 2650278 | 18426 | -5.284 | -0.1846 | Yes | ||
| 156 | DNAJB1 | 1090041 | 18430 | -5.350 | -0.1495 | Yes | ||
| 157 | CDC42EP3 | 2480138 | 18467 | -5.715 | -0.1137 | Yes | ||
| 158 | DEF6 | 840593 | 18489 | -6.000 | -0.0752 | Yes | ||
| 159 | KCNH2 | 1170451 | 18497 | -6.062 | -0.0356 | Yes | ||
| 160 | PPP1R9B | 3130619 | 18523 | -6.362 | 0.0050 | Yes |

