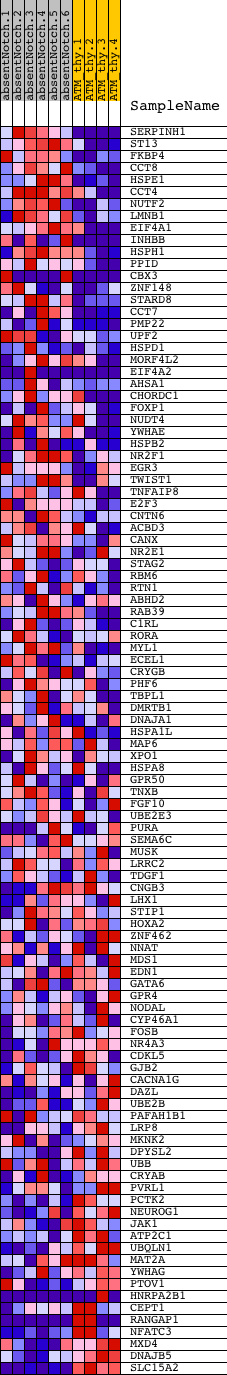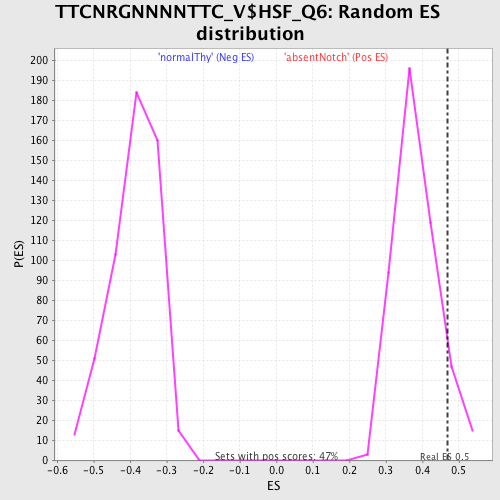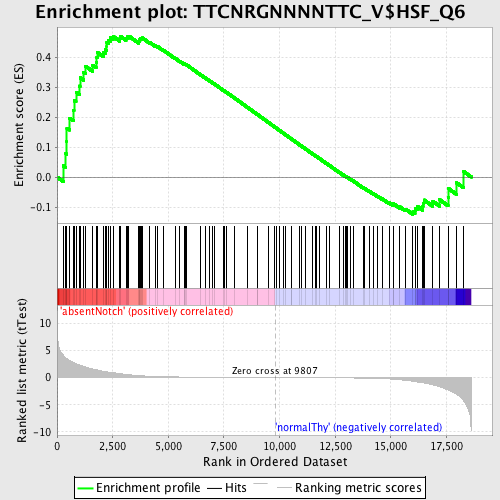
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | absentNotch |
| GeneSet | TTCNRGNNNNTTC_V$HSF_Q6 |
| Enrichment Score (ES) | 0.46995398 |
| Normalized Enrichment Score (NES) | 1.2271367 |
| Nominal p-value | 0.080168776 |
| FDR q-value | 0.9745417 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SERPINH1 | 6130014 | 276 | 4.115 | 0.0376 | Yes | ||
| 2 | ST13 | 3850369 | 390 | 3.616 | 0.0777 | Yes | ||
| 3 | FKBP4 | 870035 | 434 | 3.492 | 0.1199 | Yes | ||
| 4 | CCT8 | 6220019 | 441 | 3.449 | 0.1636 | Yes | ||
| 5 | HSPE1 | 3170438 | 569 | 3.119 | 0.1965 | Yes | ||
| 6 | CCT4 | 6380161 | 744 | 2.730 | 0.2220 | Yes | ||
| 7 | NUTF2 | 1410471 5720050 | 761 | 2.707 | 0.2557 | Yes | ||
| 8 | LMNB1 | 5890292 6020008 | 851 | 2.519 | 0.2830 | Yes | ||
| 9 | EIF4A1 | 1990341 2810300 | 1010 | 2.286 | 0.3037 | Yes | ||
| 10 | INHBB | 2680092 | 1032 | 2.255 | 0.3313 | Yes | ||
| 11 | HSPH1 | 1690017 | 1186 | 2.075 | 0.3495 | Yes | ||
| 12 | PPID | 2680707 | 1283 | 1.961 | 0.3694 | Yes | ||
| 13 | CBX3 | 1780592 3940008 | 1593 | 1.581 | 0.3728 | Yes | ||
| 14 | ZNF148 | 6420403 | 1754 | 1.441 | 0.3826 | Yes | ||
| 15 | STARD8 | 1780047 | 1785 | 1.414 | 0.3990 | Yes | ||
| 16 | CCT7 | 6660091 | 1807 | 1.392 | 0.4156 | Yes | ||
| 17 | PMP22 | 4010239 6550072 | 2087 | 1.152 | 0.4153 | Yes | ||
| 18 | UPF2 | 4810471 | 2162 | 1.087 | 0.4251 | Yes | ||
| 19 | HSPD1 | 60097 2030451 | 2231 | 1.033 | 0.4347 | Yes | ||
| 20 | MORF4L2 | 6450133 | 2235 | 1.032 | 0.4477 | Yes | ||
| 21 | EIF4A2 | 1170494 1740711 2850504 | 2303 | 1.005 | 0.4569 | Yes | ||
| 22 | AHSA1 | 2030520 | 2391 | 0.974 | 0.4646 | Yes | ||
| 23 | CHORDC1 | 6650577 | 2518 | 0.894 | 0.4692 | Yes | ||
| 24 | FOXP1 | 6510433 | 2822 | 0.711 | 0.4619 | Yes | ||
| 25 | NUDT4 | 6760689 | 2840 | 0.702 | 0.4700 | Yes | ||
| 26 | YWHAE | 5310435 | 3098 | 0.572 | 0.4634 | No | ||
| 27 | HSPB2 | 1990110 | 3153 | 0.546 | 0.4674 | No | ||
| 28 | NR2F1 | 4120528 360402 | 3230 | 0.514 | 0.4699 | No | ||
| 29 | EGR3 | 6940128 | 3667 | 0.363 | 0.4510 | No | ||
| 30 | TWIST1 | 5130619 | 3680 | 0.358 | 0.4549 | No | ||
| 31 | TNFAIP8 | 2760551 | 3716 | 0.346 | 0.4574 | No | ||
| 32 | E2F3 | 50162 460180 | 3726 | 0.343 | 0.4613 | No | ||
| 33 | CNTN6 | 2630711 | 3804 | 0.322 | 0.4613 | No | ||
| 34 | ACBD3 | 2760170 | 3826 | 0.317 | 0.4642 | No | ||
| 35 | CANX | 4760402 | 4161 | 0.244 | 0.4492 | No | ||
| 36 | NR2E1 | 4730288 | 4420 | 0.202 | 0.4379 | No | ||
| 37 | STAG2 | 4540132 | 4512 | 0.189 | 0.4354 | No | ||
| 38 | RBM6 | 840279 3840563 4780129 6020446 | 4776 | 0.156 | 0.4232 | No | ||
| 39 | RTN1 | 4150452 4850176 | 5323 | 0.112 | 0.3951 | No | ||
| 40 | ABHD2 | 4850504 6840010 | 5511 | 0.101 | 0.3863 | No | ||
| 41 | RAB39 | 430524 | 5710 | 0.090 | 0.3767 | No | ||
| 42 | C1RL | 1690142 6650092 | 5711 | 0.090 | 0.3779 | No | ||
| 43 | RORA | 380164 5220215 | 5758 | 0.089 | 0.3765 | No | ||
| 44 | MYL1 | 2450707 | 5807 | 0.086 | 0.3750 | No | ||
| 45 | ECEL1 | 5700504 | 6455 | 0.063 | 0.3409 | No | ||
| 46 | CRYGB | 2340593 | 6660 | 0.057 | 0.3306 | No | ||
| 47 | PHF6 | 3120632 3990041 | 6828 | 0.052 | 0.3223 | No | ||
| 48 | TBPL1 | 3610332 | 6996 | 0.048 | 0.3138 | No | ||
| 49 | DMRTB1 | 2470746 | 7070 | 0.046 | 0.3105 | No | ||
| 50 | DNAJA1 | 1660010 2320358 6840136 | 7463 | 0.039 | 0.2898 | No | ||
| 51 | HSPA1L | 4010538 | 7537 | 0.037 | 0.2863 | No | ||
| 52 | MAP6 | 6860195 | 7600 | 0.036 | 0.2834 | No | ||
| 53 | XPO1 | 540707 | 7976 | 0.029 | 0.2636 | No | ||
| 54 | HSPA8 | 2120315 1050132 6550746 | 8542 | 0.020 | 0.2333 | No | ||
| 55 | GPR50 | 6420112 | 9013 | 0.012 | 0.2080 | No | ||
| 56 | TNXB | 630592 3830020 5360497 7000673 | 9022 | 0.012 | 0.2078 | No | ||
| 57 | FGF10 | 730458 840706 | 9508 | 0.005 | 0.1816 | No | ||
| 58 | UBE2E3 | 2030347 3840471 | 9780 | 0.000 | 0.1670 | No | ||
| 59 | PURA | 3870156 | 9881 | -0.001 | 0.1616 | No | ||
| 60 | SEMA6C | 460458 1580110 | 9980 | -0.003 | 0.1564 | No | ||
| 61 | MUSK | 1190113 | 10174 | -0.006 | 0.1460 | No | ||
| 62 | LRRC2 | 3130400 | 10267 | -0.007 | 0.1411 | No | ||
| 63 | TDGF1 | 1690338 6760070 | 10545 | -0.011 | 0.1263 | No | ||
| 64 | CNGB3 | 2340408 | 10546 | -0.011 | 0.1265 | No | ||
| 65 | LHX1 | 1690100 | 10912 | -0.017 | 0.1070 | No | ||
| 66 | STIP1 | 6450390 | 10994 | -0.019 | 0.1028 | No | ||
| 67 | HOXA2 | 2120121 | 11148 | -0.021 | 0.0948 | No | ||
| 68 | ZNF462 | 6900100 | 11469 | -0.027 | 0.0779 | No | ||
| 69 | NNAT | 4920053 5130253 | 11606 | -0.030 | 0.0709 | No | ||
| 70 | MDS1 | 6400072 | 11641 | -0.031 | 0.0695 | No | ||
| 71 | EDN1 | 1770047 | 11782 | -0.033 | 0.0623 | No | ||
| 72 | GATA6 | 1770010 | 12104 | -0.041 | 0.0455 | No | ||
| 73 | GPR4 | 2060358 | 12254 | -0.044 | 0.0380 | No | ||
| 74 | NODAL | 2030286 7000736 | 12673 | -0.055 | 0.0162 | No | ||
| 75 | CYP46A1 | 2470332 | 12850 | -0.061 | 0.0074 | No | ||
| 76 | FOSB | 1940142 | 12955 | -0.064 | 0.0026 | No | ||
| 77 | NR4A3 | 2900021 5860095 5910039 | 13009 | -0.066 | 0.0006 | No | ||
| 78 | CDKL5 | 2450070 | 13048 | -0.067 | -0.0006 | No | ||
| 79 | GJB2 | 5900609 6200487 | 13175 | -0.073 | -0.0065 | No | ||
| 80 | CACNA1G | 3060161 4810706 5550722 | 13305 | -0.078 | -0.0124 | No | ||
| 81 | DAZL | 4050082 | 13759 | -0.102 | -0.0356 | No | ||
| 82 | UBE2B | 4780497 | 13803 | -0.105 | -0.0366 | No | ||
| 83 | PAFAH1B1 | 4230333 6420121 6450066 | 14030 | -0.123 | -0.0472 | No | ||
| 84 | LRP8 | 3610746 5360035 | 14209 | -0.138 | -0.0551 | No | ||
| 85 | MKNK2 | 1240020 5270092 | 14407 | -0.157 | -0.0637 | No | ||
| 86 | DPYSL2 | 2100427 3130112 5700324 | 14617 | -0.185 | -0.0726 | No | ||
| 87 | UBB | 4810138 | 14920 | -0.242 | -0.0859 | No | ||
| 88 | CRYAB | 4810619 | 15117 | -0.296 | -0.0927 | No | ||
| 89 | PVRL1 | 2570167 5220519 5890500 3190309 | 15123 | -0.296 | -0.0892 | No | ||
| 90 | PCTK2 | 5700673 | 15400 | -0.390 | -0.0991 | No | ||
| 91 | NEUROG1 | 380341 | 15656 | -0.499 | -0.1065 | No | ||
| 92 | JAK1 | 5910746 | 15995 | -0.673 | -0.1162 | No | ||
| 93 | ATP2C1 | 2630446 6520253 | 16088 | -0.720 | -0.1119 | No | ||
| 94 | UBQLN1 | 110280 730376 2470133 6590594 | 16106 | -0.736 | -0.1035 | No | ||
| 95 | MAT2A | 4070026 4730079 6020280 | 16205 | -0.802 | -0.0985 | No | ||
| 96 | YWHAG | 3780341 | 16437 | -0.964 | -0.0987 | No | ||
| 97 | PTOV1 | 4230025 | 16449 | -0.974 | -0.0869 | No | ||
| 98 | HNRPA2B1 | 1240619 6200161 6200494 6980408 | 16491 | -0.998 | -0.0764 | No | ||
| 99 | CEPT1 | 1940047 | 16886 | -1.339 | -0.0805 | No | ||
| 100 | RANGAP1 | 2320593 6650601 | 17194 | -1.699 | -0.0754 | No | ||
| 101 | NFATC3 | 5050520 | 17575 | -2.268 | -0.0670 | No | ||
| 102 | MXD4 | 4810113 | 17601 | -2.318 | -0.0388 | No | ||
| 103 | DNAJB5 | 2900215 | 17957 | -3.044 | -0.0191 | No | ||
| 104 | SLC15A2 | 1780538 | 18277 | -4.282 | 0.0183 | No |